Protein interaction prediction method
A prediction method, a protein technology, applied in the field of bioinformatics, can solve problems with selection bias, unable to cover NIPs, etc., and achieve the effects of good robustness, easy generalization, and good predictive performance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0029] The present invention will be further described below in conjunction with the accompanying drawings and specific embodiments. It should be understood that these examples are only used to illustrate the present invention and are not intended to limit the scope of the present invention. The operating methods not indicated in the following examples are generally in accordance with conventional conditions, or in accordance with the conditions suggested by the manufacturer.
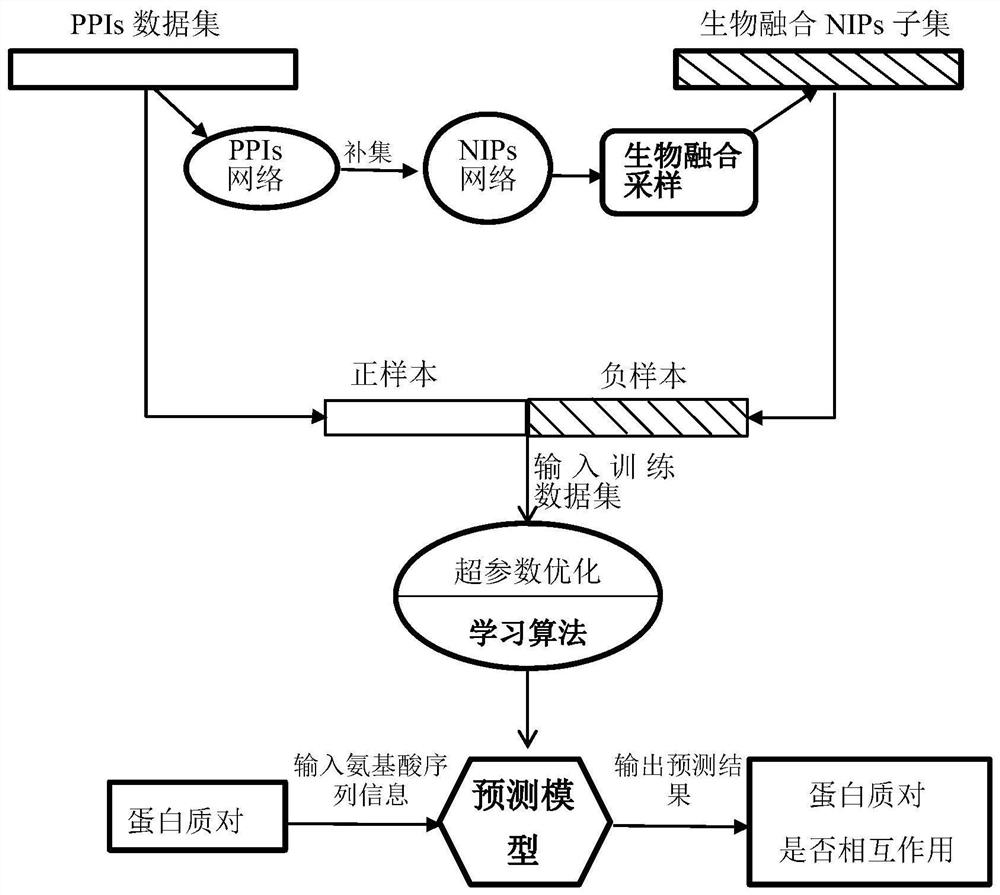
[0030] The protein interaction prediction method of the present invention based on the sampling strategy of non-interacting protein pairs fused with biological semantics is as follows: figure 1 Shown, specifically, include the steps:
[0031] (A) Yeast PPIs data were obtained from the S. cerevisiae core subset (“Scere20080708.txt”) in the DIP database (Lukasz, Salwinski et al., Nucleic Acids Research, suppl_1 (2004): suppl_1.). The original yeast PPIs data was first clustered and analyzed using the CD...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com