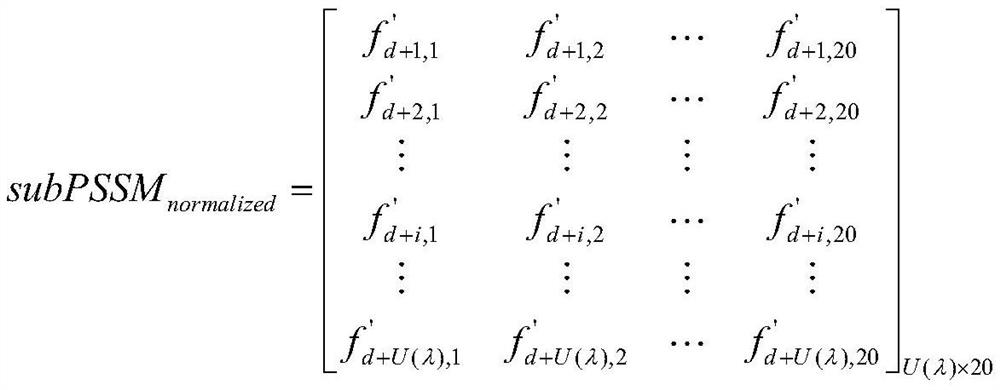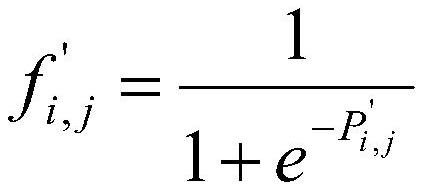DNA binding protein prediction method based on local evolution information
A protein-binding and prediction method technology, applied in the field of bioinformatics DNA-binding protein prediction, can solve the problems of weakening the influence of global information on the model contribution, large model scale redundancy, insufficient model prediction efficiency, etc., to achieve training efficiency and prediction. Efficiency improvement, model accuracy improvement, model efficiency improvement effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
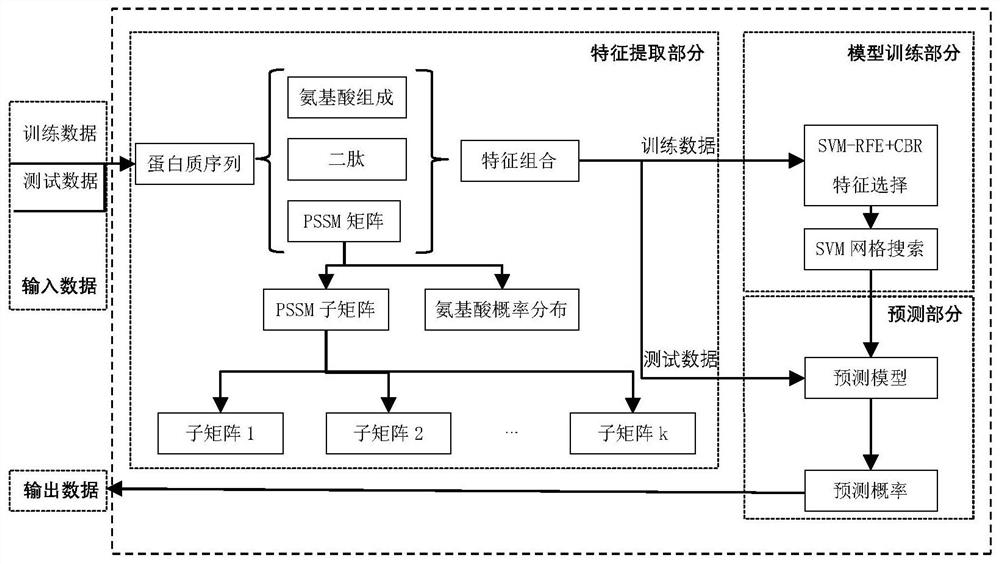
[0050] like figure 1 As shown, in this embodiment, a DNA-binding protein prediction method based on local evolution information includes the following steps:
[0051] Step 1: Feature Extraction
[0052] Given a protein sequence S, denoted as S 1 S 2 S 3 …S L , where S i (1≤i≤L) is the amino acid (residue) appearing at position i, and L is the length of the protein sequence S. Use PSI-BLAST to obtain protein evolution information PSSM. The PSSM matrix is a matrix of L×20 (L rows and 20 columns), and its format is as follows:
[0053]
[0054] where L is the length of the original protein sequence, p i,j (i=1, 2, 3...L, j=1, 2, 3...20) is the probability score of the amino acid at position i evolving into position j in the protein sequence.
[0055] By dividing the PSSM into k equal PSSM matrices by row, the sub-matrix formula is obtained as:
[0056]
[0057] Among them, d=(λ-1)×U(λ), indicating that the starting sequence position of each sub-matrix is in th...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com