Recombinant herpes simplex virus and method for preparing same
A herpes simplex virus and genome technology, applied in biochemical equipment and methods, viruses, virus/bacteriophage, etc., can solve the problems of virus replication limitation, difficult genetic manipulation, and low therapeutic efficacy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
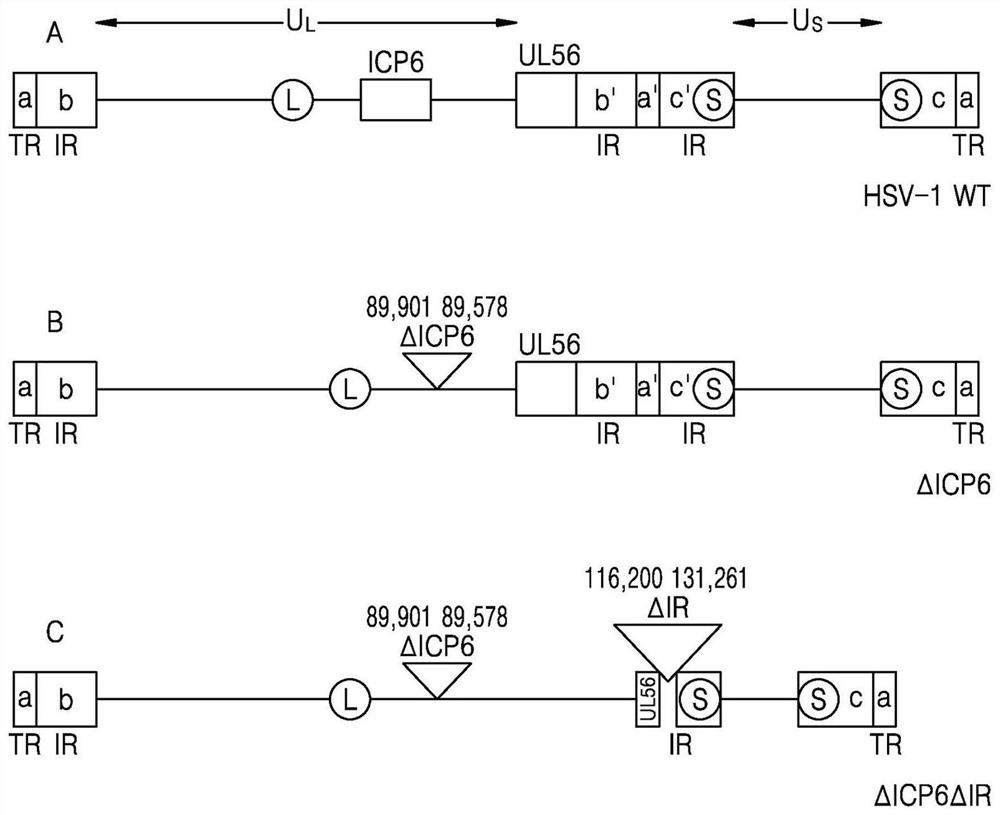
[0073] Embodiment 1: Make wherein delete the recombinant HSV-1 of ICP6 and IR region (ΔICP6ΔIR HSV-1)
[0074] figure 1 A is a schematic diagram of the genome of HSV-1. The HSV-1 genome is mainly divided into a long unique (Unique Long: UL) region and a short unique (Unique short: US) region. In the UL region, a single origin of replication and a and b consisting of terminal repeats (Terminal repeat: TR) and IR are located at 5', while a and b in the reverse direction are located at 3'. In the US region, two replication origins and the c sequence in the reverse direction are located at the 5', the c sequence in the forward direction is repeated at the back, and the a sequence is repeated once at the 3' end.
[0075] First, a ΔICP6 HSV-1 virus was produced in which a part of the ICP6 region was deleted from the genome sequence of the wild-type HSV-1 WT virus. Nucleotides 86,364 to 89,777 in the wild-type HSV-1 genome sequence are sequences encoding the ICP6 gene. Nucleotide...
Embodiment 2
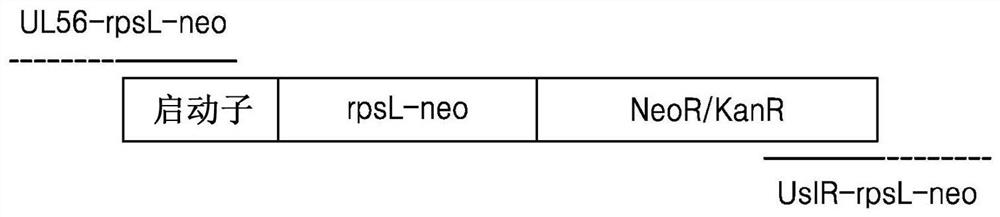
[0078] Example 2: Production of ΔICP6-ΔIR-rpsL-neo into which foreign gene rpsL-neo is inserted
[0079] A virus was produced by inserting rpsL-neo gene 1319 bp as a foreign gene into the ΔICP6ΔIR HSV-1 of Example 1 in which both the ICP6 gene and the IR gene were deleted, and it was verified whether the foreign gene was inserted.
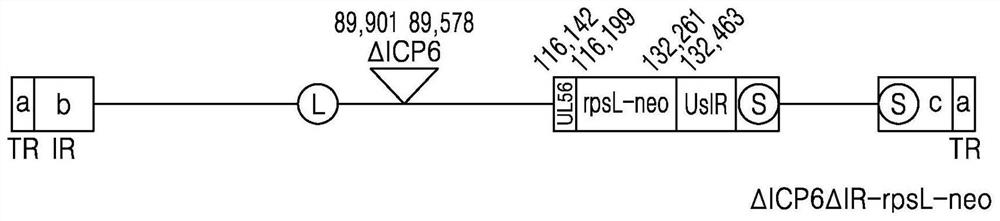
[0080] Specifically, as shown in Table 1 below, the UL56-rpsL-neo primer with a sequence complementary to UL56 and rpsL-neo and the UsIR-rpsL-neo primer with a sequence complementary to UsIR and rpsL-neo were made with 200 ng of rpsL- The neo gene was used as a template, and PCR was performed using 10 pmol of UL56-rpsL-neo primer, UsIR-rpsL-neo primer and 2X green master mix (thermo scientific) reaction enzyme. The PCR product was transformed into DH10b Escherichia coli together with the ΔICP6 total vector of Example 1 to induce homologous recombination (homologous recombination), thereby producing ΔICP6-ΔIR-rpsL-neo expressing rpsL-neo.
[0081] ...
Embodiment 3
[0092] Example 3: Making ΔICP6ΔIR virus
[0093] The ΔICP6ΔIR virus was prepared by removing the foreign gene rpsL-neo from the ΔICP6-ΔIR-rpsL-neo prepared in Example 2 above.
[0094] Specifically, after preparing the primers in Table 3 below, a double-stranded oligomer bound with a complementary sequence was prepared by performing a slow cooling reduction method (primer annealing). The produced oligomer was transformed into DH10b Escherichia coli together with the ΔICP6-ΔIR-rpsL-neo total vector of Example 2 to induce homologous recombination, thereby producing dICP6-dIR.
[0095] 【table 3】
[0096]
[0097] In order to confirm whether the ΔICP6ΔIR virus from which the foreign gene was removed was successfully produced, PCR was carried out using the above-mentioned PCR primers. Using 200ng as a template, after adding 2X green master mix (thermo scientific) reaction enzyme to 10pmol of UL55 S primer and ΔIR AS primer, perform PCR in a PCR machine (Bio-rad) according to t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com