MiRNA-disease association prediction method and system based on tensor decomposition
A technology of tensor decomposition and prediction method, applied in the field of bioinformatics, to achieve the effect of good prediction performance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0068] The scheme of the present invention will be further described below in conjunction with examples and accompanying drawings.
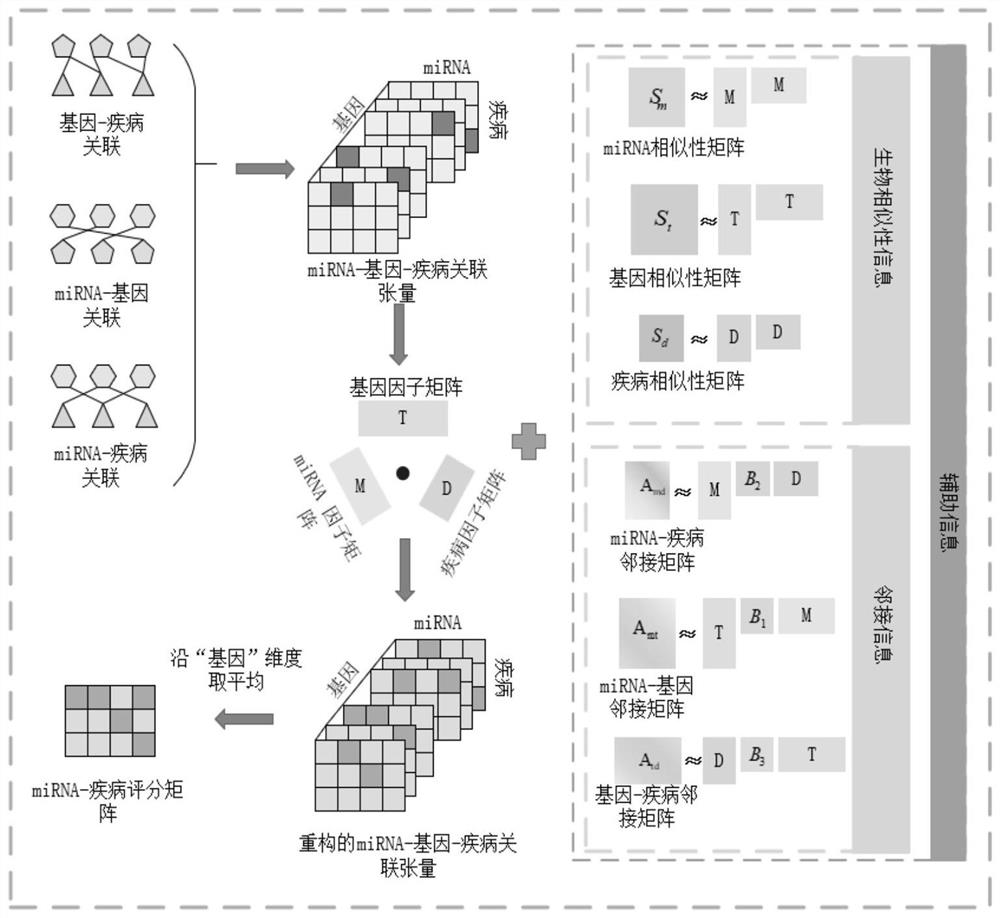
[0069] Such as figure 1 As shown, a tensor decomposition-based miRNA-disease association prediction method includes the following steps:
[0070] Step 1: Construct miRNA-gene-disease association tensor based on known miRNA-disease association data, miRNA-gene association data and gene-disease association data;
[0071] Download the miRNA-disease association data from the HMDD database, download the miRNA-gene association data from the miRNATarBase database, and download the gene-disease association data from DisGeNet.
[0072] Step 1 specifically includes the following steps:
[0073] Step 11: Construct the miRNA-gene-disease association tensor:
[0074] Merge the known miRNA-disease association data, miRNA-gene association data and gene-disease association data to construct the association dataset . Associations are modeled by a rank 3 tenso...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com