Drop-off ddPCR method and kit for quantitatively detecting NPM1 gene mutation
A quantitative detection and kit technology, which is used in DNA/RNA fragmentation, microbial determination/inspection, biochemical equipment and methods, etc., and can solve problems such as low sensitivity, single mutation type, and inability to perform absolute quantification by real-time fluorescent quantitative PCR. , to achieve high sensitivity and specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0045] Taking the bone marrow samples of patients with NPM1 gene type A mutation as an example, the detection method is described in detail.
[0046] 1. Isolation of bone marrow mononuclear cells and extraction of DNA
[0047] (1) Add 5ml red blood cell lysate to the bone marrow sample, mix well, and let stand for 1min;
[0048] (2) Centrifuge at 12000×rpm for 20sec, discard the supernatant and keep the precipitate;
[0049] (3) Add 1ml Trizol and mix well by pipetting;
[0050](4) Add 200 μl of chloroform, vortex for 20 sec, let stand for 10 min, centrifuge at 12000×rpm for 1 min, and discard the supernatant;
[0051] (5) After centrifuging again at 12000×rpm, discard the supernatant and keep the precipitate, and the white precipitate of DNA at the bottom of the tube can be seen;
[0052] (6) Add 1ml of trisodium citrate to the tube, flick or invert to mix and rinse the precipitate, let stand for 20-30min, centrifuge at 12000×rpm for 5min, and discard the supernatant;
[...
Embodiment 2
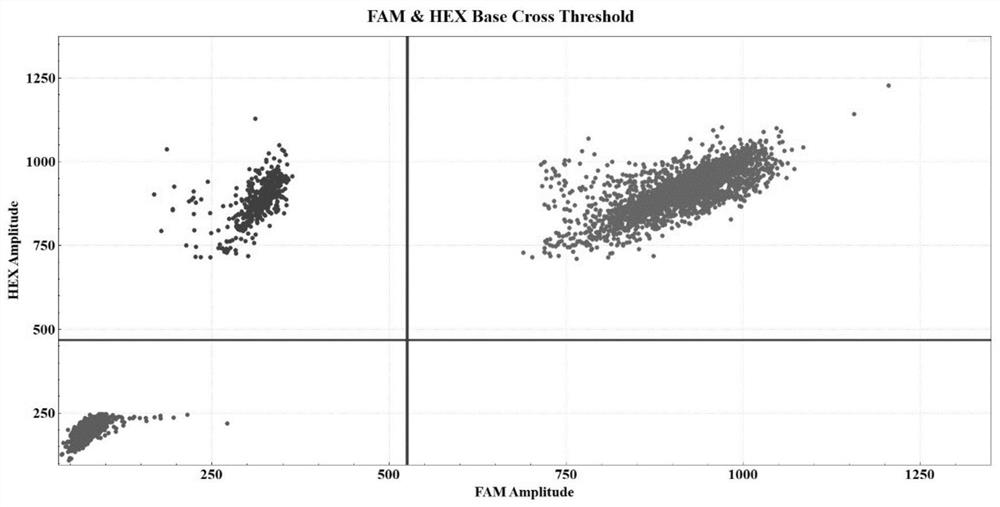
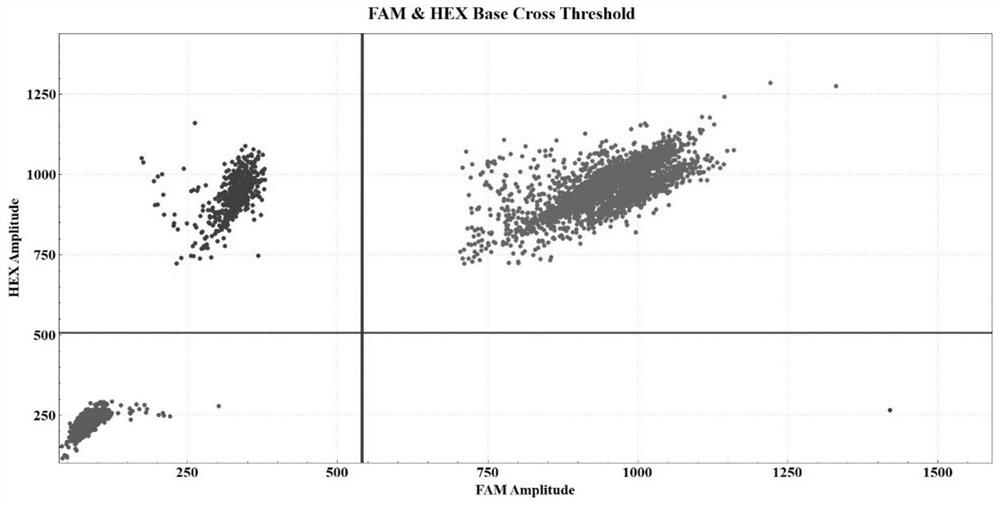
[0074] Taking the bone marrow sample of a patient with a D-type mutation of the NPM1 gene as an example, the specific detection process is the same as that in Example 1. After data analysis, it can be obtained as figure 2 The experimental results and related data. In 250ng sample DNA, the concentration of FAM is 1660.10copies / μl, the concentration of VIC is 1921.90copies / μl, the calculated concentration of NPM1 D-type mutation template is 261.80copies / μl, and the allele frequency of NPM1 is 13.62%.
Embodiment 3
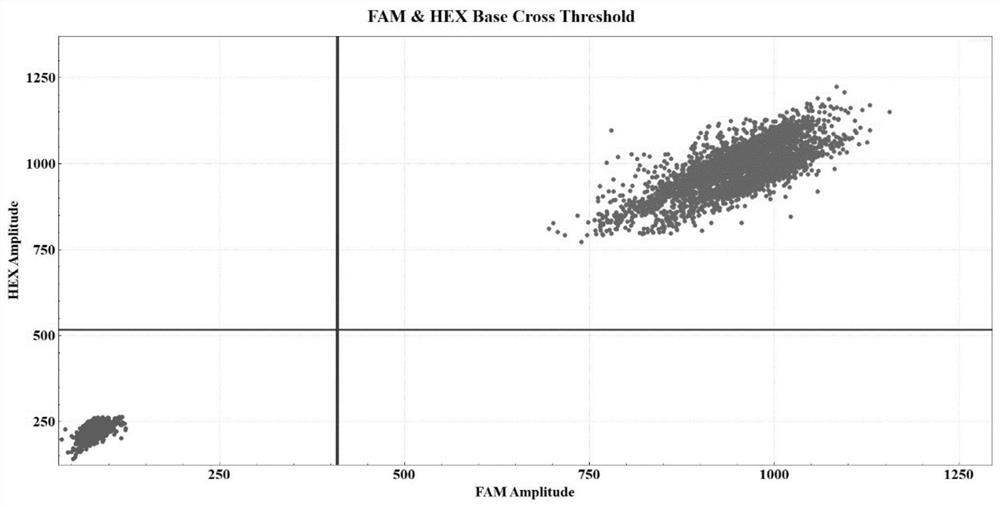
[0076] Taking the bone marrow sample of a patient with negative NPM1 gene mutation as an example, the specific detection process is the same as that in Example 1. After data analysis, it can be obtained as image 3 The experimental results and related data. In 250ng sample DNA, the concentration of FAM was 2078.37copies / μl, the concentration of VIC was 2078.37copies / μl, and the calculated concentration of NPM1 mutation template was 0copies / μl, which was lower than the detection limit (22.78copies / μl), so the patient’s NPM1 gene mutation was negative.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com