Methods of treating cancer with farnesyltransferase inhibitors
An inhibitor, cancer technology, applied in the field of cancer therapy, can solve problems such as unmet sexual needs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
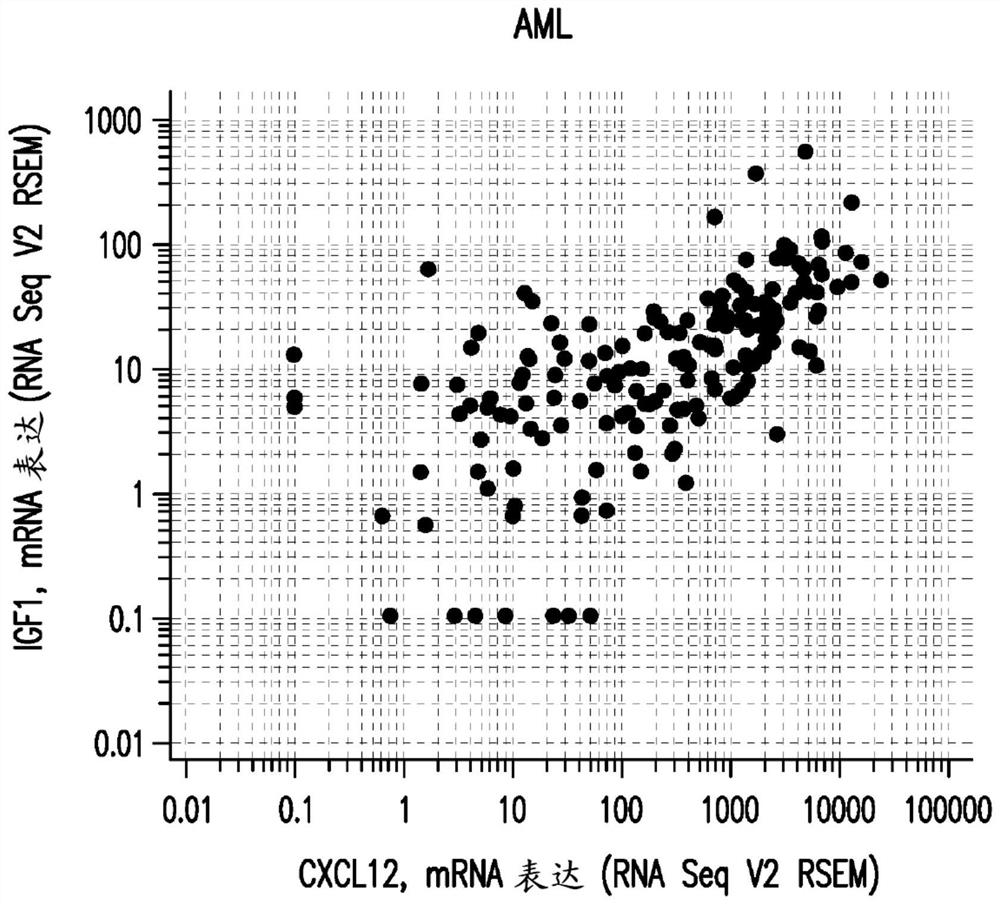
[0461] Example 1: Crosstalk between the IGF1 and CXCL12 pathways defines targeted responses to the farnesyltransferase inhibitor tipifarnib in AML and PTCL patients
[0462] Results were analyzed using RNA-Seq and Affymetrix U133A microarray analysis of tumor samples from 71 patients enrolled in the Tipifarnib trials (CTEP-20, KO-TIP-002, KO-TIP-004). Gene Expression Profile (GEP) data, and complement the analysis of mRNA expression in datasets from cBioportal for Cancer Genomics. Clinical trial information: NCT00027872, NCT02464228, NCT02807272.
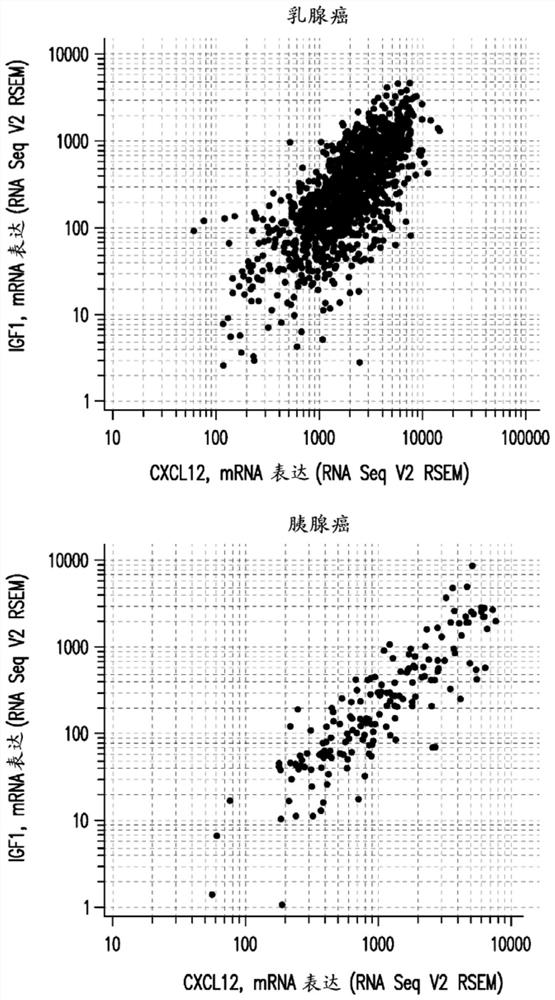
[0463] The pathology of tumor CXCL12 overexpression was explored by examining the gene expression signature (available at cBioportal (TCGA, Provisional)) of 8,401 cancer patients in 25 studies. In 19 of these studies, the expression of IGF1 and CXCL12 genes A highly significant correlation was present. These 25 studies are detailed in Table 1 below. The highly significant correlation between IGF1 and CXCL12 gene expression in breas...
Embodiment 2
[0476] Example 2: Identification of clinical and molecular biomarkers associated with clinical benefit of tipifarnib in advanced pancreatic cancer.
[0477] as table 1 and figure 1 As seen in , patients with pancreatic cancer showed a high correlation between the CXCL12 and IGF1 genes and higher than other tumor types for which tipifarnib activity has been observed. Pancreatic tumors also express very high levels of IGFB7. IGFBP7 was found to be co-expressed in these tumors with IGF1 (p=0.643, p<0.0001, N=179) and CXCL12 (p=0.617, p<0.0001, N=173).
[0478] In a randomized, double-blind, placebo-controlled study of gemcitabine + tipifarnib versus gemcitabine + placebo in patients with advanced pancreatic adenocarcinoma not previously treated with systemic Tipifarnib (R115777) was administered at a dose of 200 mg (Study INT-17). Gemcitabine was administered intravenously at a dose of 1,000 mg / m(2) 7 times a week for 8 weeks, then 3 times a week every 4 weeks.
[0479] 686 p...
Embodiment 3
[0488] Example 3: Low KRAS Mutant Allele Frequency Identifies Pancreatic Cancer Patients at Potential for Clinical Benefit of Tipifarnib
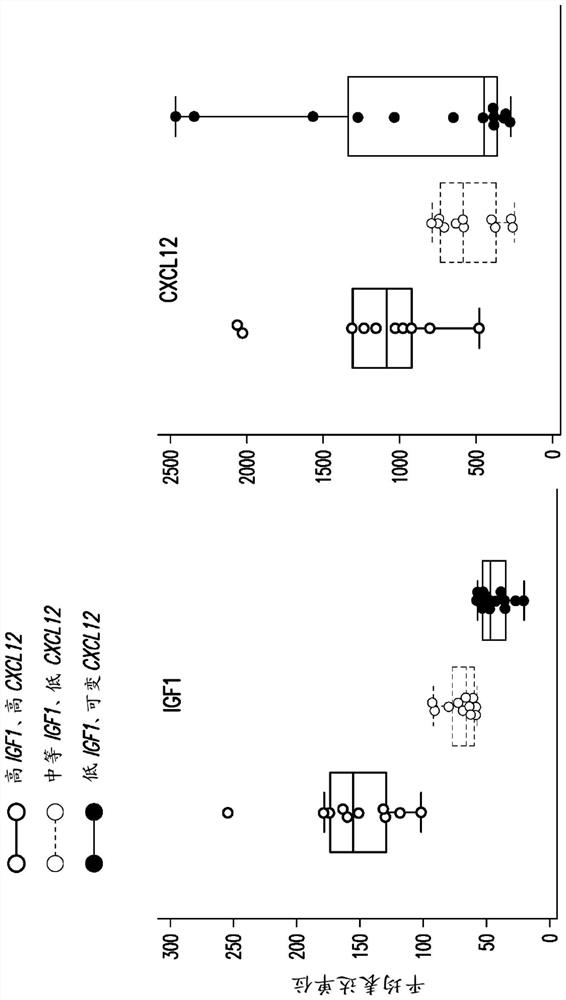
[0489] Low KRAS mutant allele frequency in pancreatic tumor samples is associated with high CXCL12 and high IGF1 gene expression. However, IGFBP7 is highly expressed in pancreatic cancer overall, but tends to increase with decreasing KRAS mutant allele frequency ( Figure 7A ). Thus, low KRAS mutant allele frequencies may identify pancreatic cancer patients with high CXCL12 who are susceptible to the clinical benefit of tipifarnib. Likewise, low KRAS mutant allele frequency identifies pancreatic cancer patients with the highest IGFBP7 expression, which blocks any potential tipifarnib resistance mediated by IGF1 ( Figure 7A ).
[0490] CXCL12 expression was also elevated in tumors with wild-type TP53 status or TP53 mutations with low allele frequency ( Figure 7B , left). The optimal cutoff for the TP53 mutant allele frequency for CXCL...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com