Whole genome typing method based on DNA methylation site genotypes
A whole-genome and typing method technology, applied in the field of whole-genome typing based on DNA methylation site genotype, can solve the problem of inaccuracy, inability to judge the change of DNA methylation site genotype, and inability to judge the environment effects and other issues to achieve the effect of improving accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0018] Next, the technical solutions in the embodiments of the present invention will be described in connection with the drawings of the embodiments of the present invention, and it is understood that the described embodiments are merely the embodiments of the present invention, not all of the embodiments. Based on the embodiments of the present invention, all other embodiments obtained by those of ordinary skill in the art are in the range of the present invention without making creative labor premise.
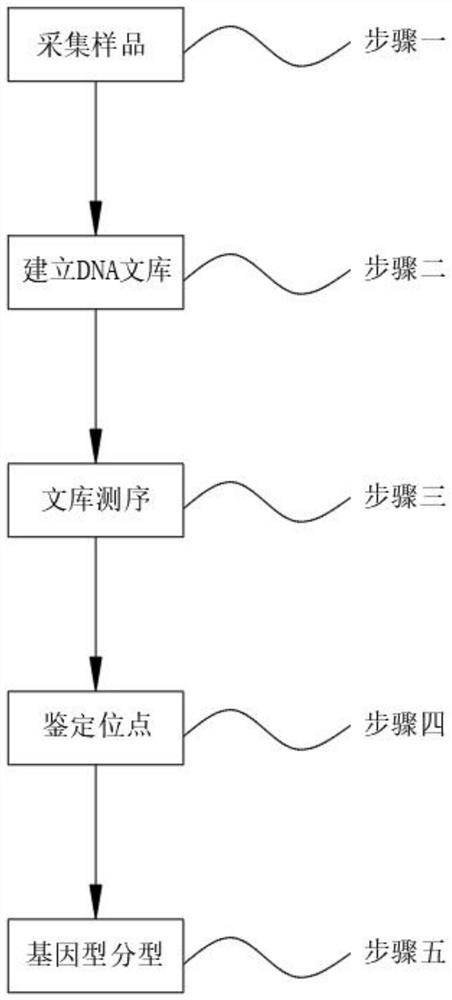
[0019] See figure 1 One embodiment of the present invention: a whole genome component method based on a DNA methylation site, comprising the steps of: Step 1, collecting a sample; step two, establish a DNA library; step three, library sequencing Step 4, identify the site; step five, genotype classification;
[0020] In the above steps, first, in the same time, in the same time, two genomic DNAs of the same tissue sample are extracted, and the same individuals are saved, and the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com