Application of SNP (Single Nucleotide Polymorphism) marker and SNP loci as well as primers, probes and detection kit of SNP marker and SNP loci
A site and marker technology, used in the determination/inspection of microorganisms, DNA/RNA fragments, recombinant DNA technology, etc., can solve the problems of low accuracy and sensitivity, and influence on the accuracy of results, and achieve low-cost urine collection. , good monitoring effect, high detection sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0127] A SNP marker for assessing the risk of graft injury and rejection after liver transplantation, the SNP markers are SNP sites rs2293840, rs2293841, rs2293843, rs163792, rs1569959, rs2250911, rs1903858, rs2293871, rs2293877, rs817363, rs22938990, 、rs704326、 rs2249102、rs2294047、rs2295499、rs2294111、rs208244、rs572496、rs2294822、 rs2295167、rs2295166、rs2295164、rs1321666、rs2295632、rs2295147、rs2974、 rs1547093、rs2295072、rs1322775、rs2255090、rs2294992、rs1022811、rs786906、 rs1738051、rs2294897、rs2294895 A combination of , rs2294886, rs2294871 and rs926587.
[0128] A kit for assessing the risk of graft injury and rejection after liver transplantation, comprising primers whose sequences are shown in SEQ ID No.1-80 in Table 1 and probes whose sequences are shown in SEQ ID Nos.81-160 Needle, ddPCR TM ProbeSupermix and ultrapure water;
[0129] The 5' ends of the probes are respectively connected with FAM fluorescent groups, and the 3' ends of the probes are labeled with NFQ and MGB.
...
Embodiment 2
[0143] Using the kit provided by the invention to assess the risk of graft injury and rejection after liver transplantation
[0144] 1. Extraction of total free DNA in urine
[0145] 10ml of urine was collected from a recipient 8 weeks after liver transplantation, and the urine was centrifuged at 3000rpm for 5min.
[0146] Take 4ml of centrifuged urine supernatant, use QIAamp Circulating NucleicAcid Kit from QIAGEN to extract total free DNA, adjust the DNA concentration to 10-30ng / ul.
[0147] 2. Prepare digital PCR reaction droplets and perform digital PCR reaction
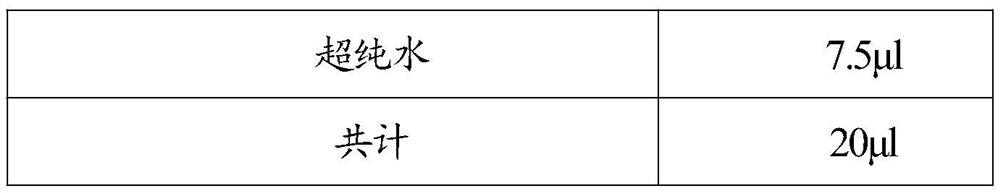
[0148] 2.1. Configure the digital PCR reaction system, prepare digital PCR reaction droplets, and perform PCR reaction; the detection system of 40 SNP sites, a total of 40 tubes, the reaction preparation is as follows:
[0149] Table 2
[0150]
[0151] Among them, the primers used to detect SNP sites include upstream and downstream primers, each 0.125 μL, a total of 0.25 μL; two probes, each 0.125 μL, a to...
Embodiment 3
[0170] Include 100 cases of liver transplant recipients diagnosed with transplant rejection and 100 cases of liver transplant recipients diagnosed with normal grafts, collect their urine according to Example 2, extract the total free DNA in the urine, and prepare digital PCR reaction microbiota. After the reaction, analyze each SNP site, calculate the GcfDNA supply-to-receiver ratio, and set 2.5% as the reference value to distinguish transplant rejection (GcfDNA supply-to-receiver ratio>2.5%) and normal graft group (GcfDNA supply to recipient ratio ≤ 2.5%) patients.
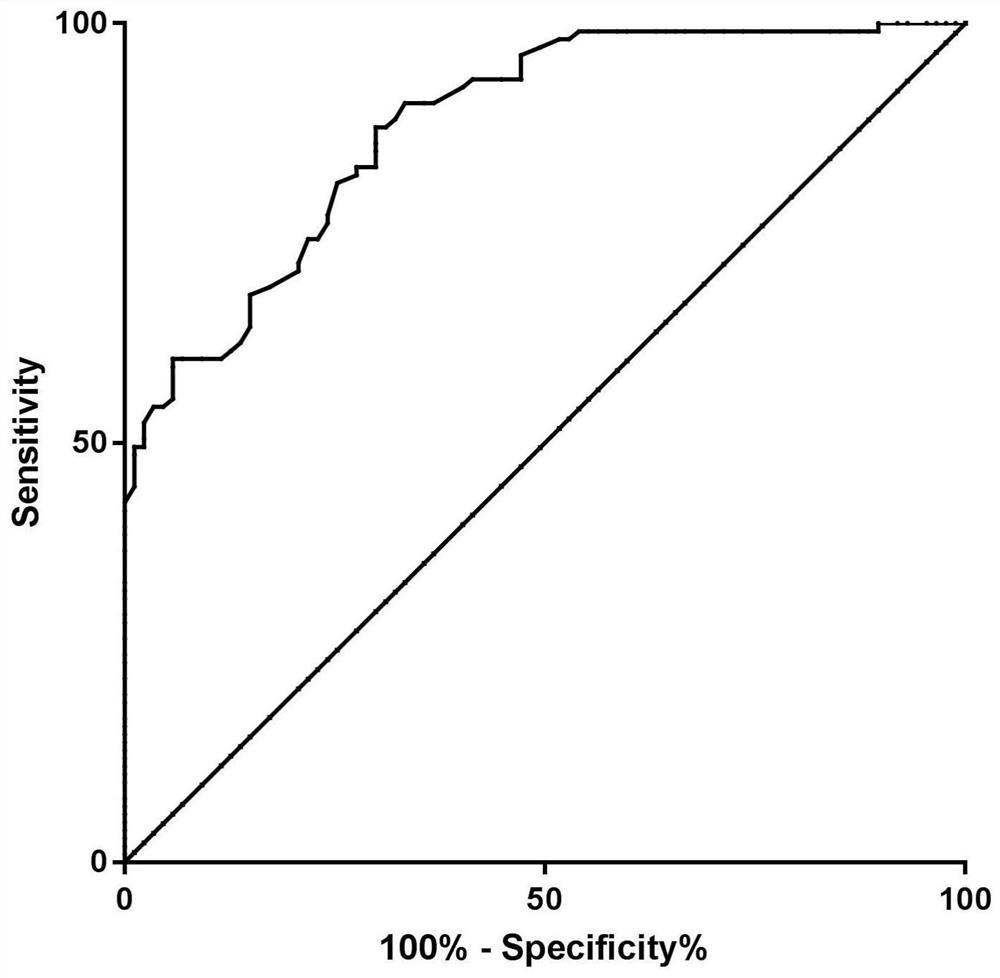
[0171] Use MedCalc software to analyze, obtain the ROC curve ( figure 1 ), AUC=0.88, sensitivity=82.86%, specificity=72.41%.
[0172] The kit of the invention can timely, accurately and specifically reflect the health status of the liver graft, and evaluate the graft injury and rejection risk after liver transplantation.
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com