Low-depth sequencing population genotype filling calculation memory optimization method
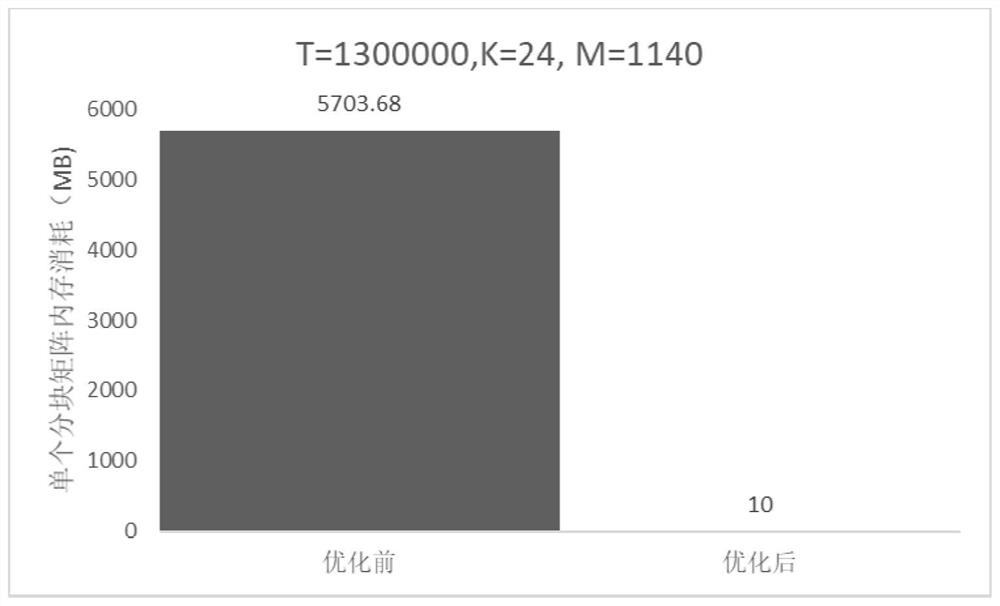
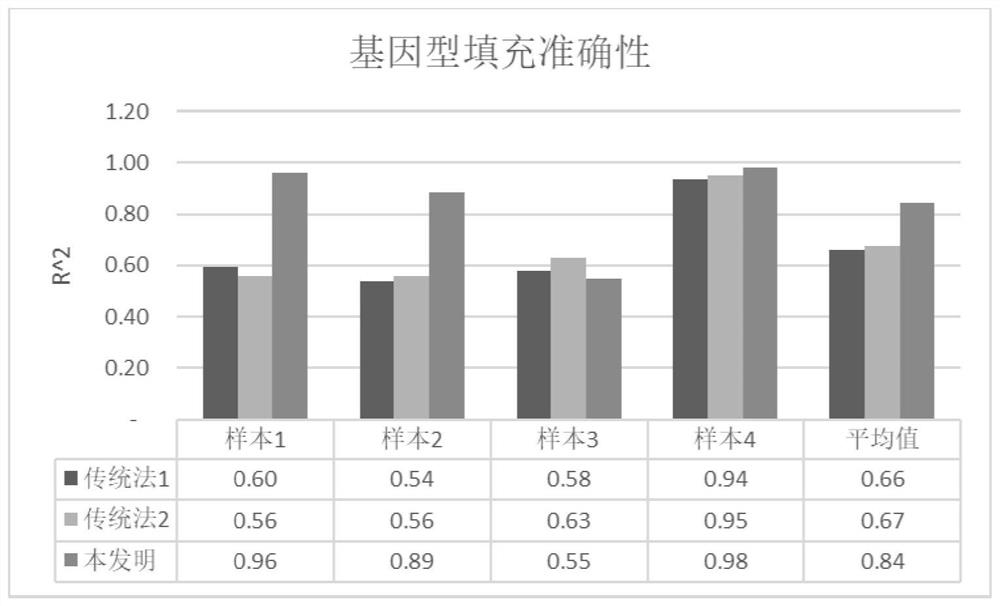
A technology of deep sequencing and optimization methods, applied in computing, genomics, program control design, etc., can solve problems such as different degrees of linkage disequilibrium, loss of filling accuracy, and failure to take into account the data dependencies of computational auxiliary variable matrices, etc. The effect of reducing memory consumption, improving efficiency, and eliminating data dependencies
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0061] The present invention will be further described in detail below in conjunction with the accompanying drawings and specific embodiments.
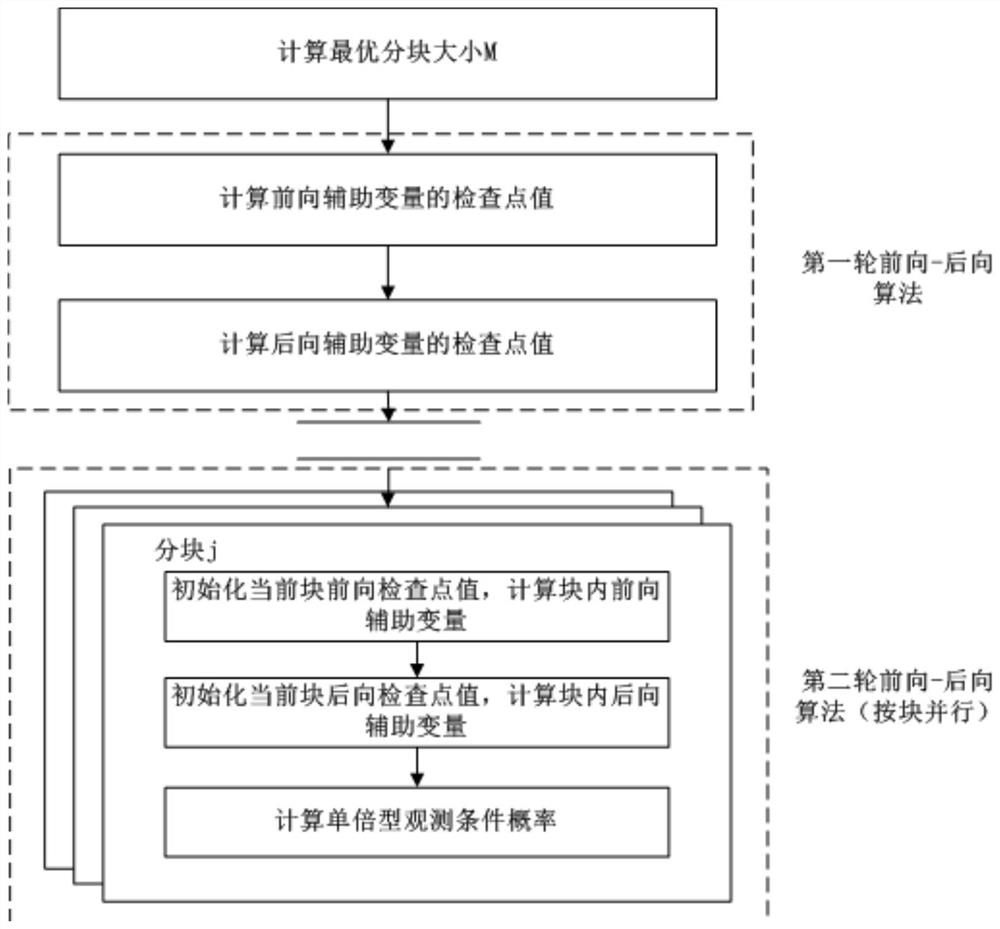
[0062] Such as figure 1 As shown, the low-depth sequencing population genotype filling calculation memory optimization method of the present invention includes:
[0063] Block the single nucleotide polymorphism site SNP, and set the checkpoint value according to the block;
[0064] According to the set checkpoint value, the forward auxiliary variable and the backward auxiliary variable are calculated by block, and the conditional probability of haplotype observation is calculated.
[0065] In a specific application example, the first round of forward auxiliary variable and backward auxiliary variable algorithm is used to determine the checkpoint; on the basis of the checkpoint value, the second round of forward auxiliary variable and backward auxiliary variable algorithm is used to calculate the block The forward auxiliary variable ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com