Nucleic acid combination and kit for colorectal cancer gene methylation detection
A colorectal cancer, methylation technology, applied in recombinant DNA technology, biochemical equipment and methods, microbial determination/inspection, etc., can solve the problem of poor sensitivity, low specificity, and single detection sensitivity of adenomas To achieve the effect of improving specificity, high detection specificity, and good comprehensive detection effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0049] Example 1. Collection and storage of feces, DNA extraction from feces
[0050] 1. Feces collection and storage
[0051] Stool sample collection steps are as follows:
[0052] (1) Defecation: Pass the feces into a squatting pan or waste box, and then take a sample immediately;
[0053] (2) Digging feces: Open the fecal DNA sample preservation tube, and use the sampling spoon on the lid to dig out a spoonful of feces (about the size of a jujube);
[0054] (3) Tighten the tube cap: Put the spoon and the stool sample back into the tube, tighten the tube cap, shake and mix for about 30 seconds.
[0055] The fecal DNA sample preservation tube used in this step can use conventional preservation tubes in the field, but it is recommended to use the feces sample collection tube of Jiangsu Kangwei Century Biotechnology Co., Ltd., the product number is CWY041. After using this product to collect feces, you can Store at 2-37°C for at least 60 days, and store at -20°C and -80°C fo...
Embodiment 2
[0070] Example 2. DNA methylation conversion
[0071] The DNA methylation conversion of stool samples can use conventional commercial kits in the field; it is recommended to use the DNA methylation conversion kit of Jiangsu Kangwei Century Biotechnology Co., Ltd., the product number is CWY105. The DNA methylation conversion steps of stool samples are as follows:
[0072] (1) Equilibrate the DNA sample that needs to be methylated to room temperature in advance; take 20 μL of the sample and add it to a new PCR tube;
[0073] (2) Add 180 μL of transformation solution to the sample, with a total volume of 200 μL, mix well and put it into the PCR instrument to start the transformation; the transformation procedure is as follows: 98°C for 10 minutes; 54°C for 1 hour; 4°C hold;
[0074] (3) Take a new centrifuge tube, add 600 μL of buffer MB, transfer the transformation liquid obtained from the reaction in step (2) into buffer MB, shake and mix;
[0075] (4) Add 20 μL of magnetic b...
Embodiment 3
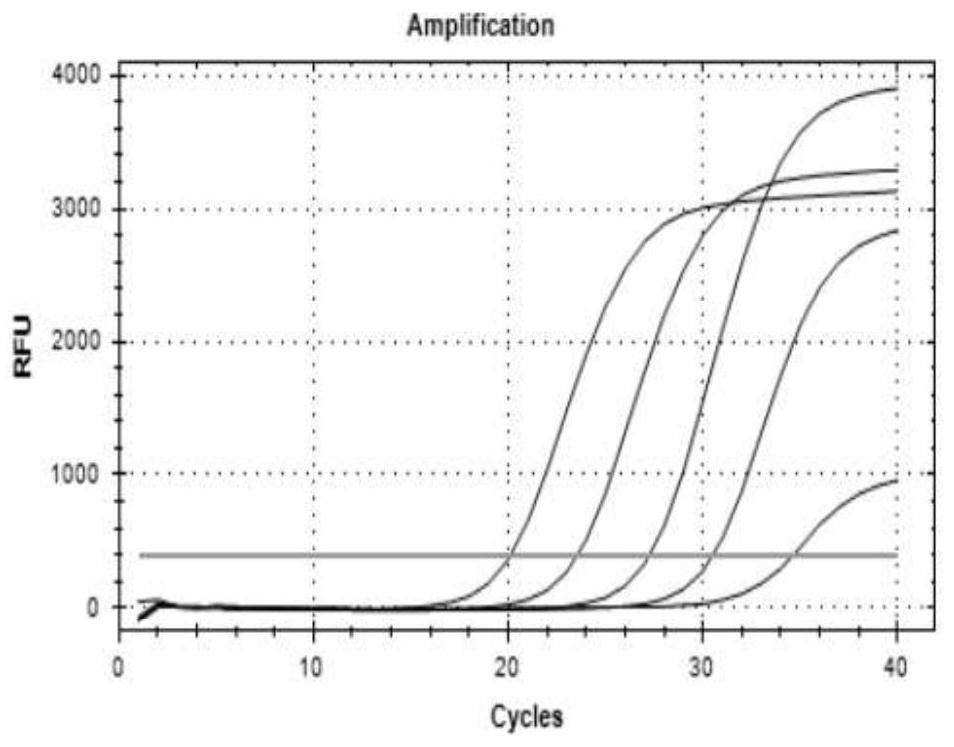
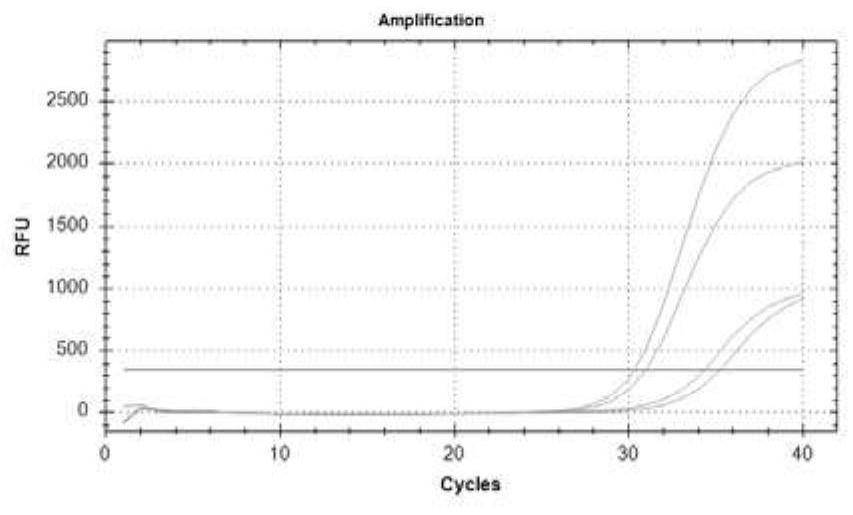
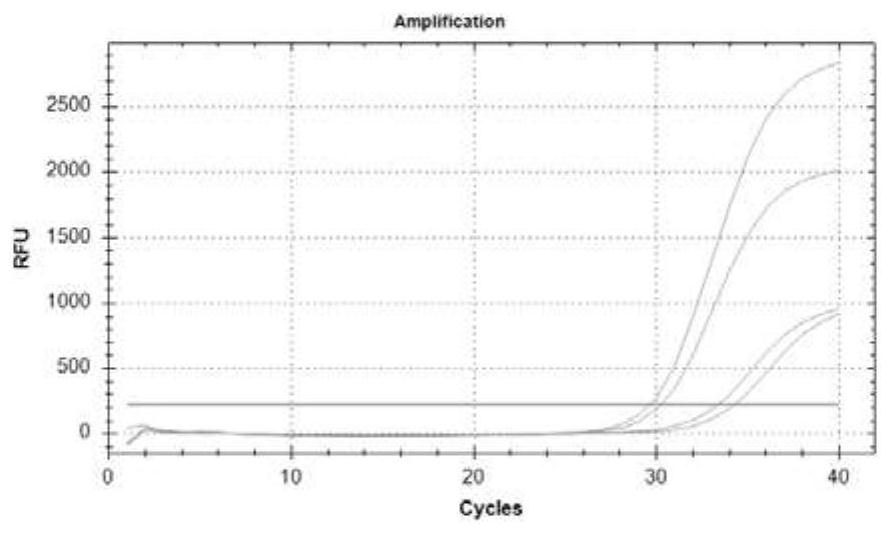
[0081]Example 3. Primer, probe design and performance evaluation for methylated gene detection
[0082] The methylation of genes mainly occurs in the promoter region, and the gene promoter region often contains multiple CpG islands. There is no clear theory that choosing which CpG island as the detection region has better diagnostic performance. In this study, by analyzing the promoter regions of three genes, two different methylated regions were obtained for each gene, named a and b regions.
[0083] 1. Design specific primer pairs and probes
[0084] Colorectal cancer-specific methylation markers septin9, SDC2, and NDRG4 were obtained for detection by reviewing literature, referring to relevant domestic and foreign methylation detection products, and searching the TCGA (The cancer genomeatlas) database for bioinformatics mining markers, etc. For markers, the promoter regions of three markers were obtained through NCBI, and after obtaining two CpG islands a and b for each ge...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
| Sensitivity | aaaaa | aaaaa |
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com