Method and device for evaluating homologous recombination defects of single sample and storage medium
A homologous recombination and single-sample technology, applied in the fields of genomics, proteomics, instruments, etc., can solve problems such as the inability to apply HRD indicators and the inability to perform accurate and effective homologous recombination defect assessment, and achieve the goal of overcoming the dependence on blood cell samples Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
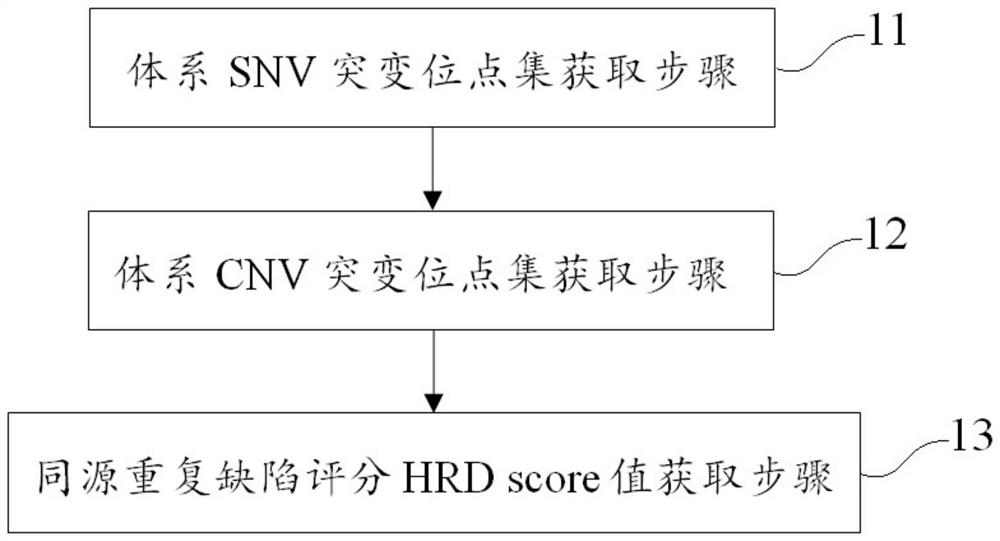
[0076] This example evaluates a method for single-sample homologous recombination deficiencies, including the following steps:
[0077] System SNV mutation site set acquisition steps: including acquiring SNV mutation sites of the tumor samples to be tested and the mutation frequency and mutation site depth of each SNV mutation site, annotating the SNV mutation sites, Tumor samples were differentiated from germline SNV mutations and systemic SNV mutations to obtain a set of systemic SNV mutation sites.
[0078] In this example, the Mutect2 module in the GATK software is used to detect the mutation frequency and depth of the mutation site in the tumor sample, and the vcf file is output, and the mutag software is used to annotate the mutation sites of the system in each database, and Screen credible systematic SNV mutation site sets according to the filtering rules provided by the interpretation. Specifically, filter out germline SNV mutations and retain systemic SNV mutations a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com