Method and device for correcting homologous recombination defect score and storage medium
A technology of homologous recombination and storage medium, applied in the fields of genomics, proteomics, instruments, etc., can solve the problems of low sensitivity and low accuracy, and achieve the effect of improving sensitivity and accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
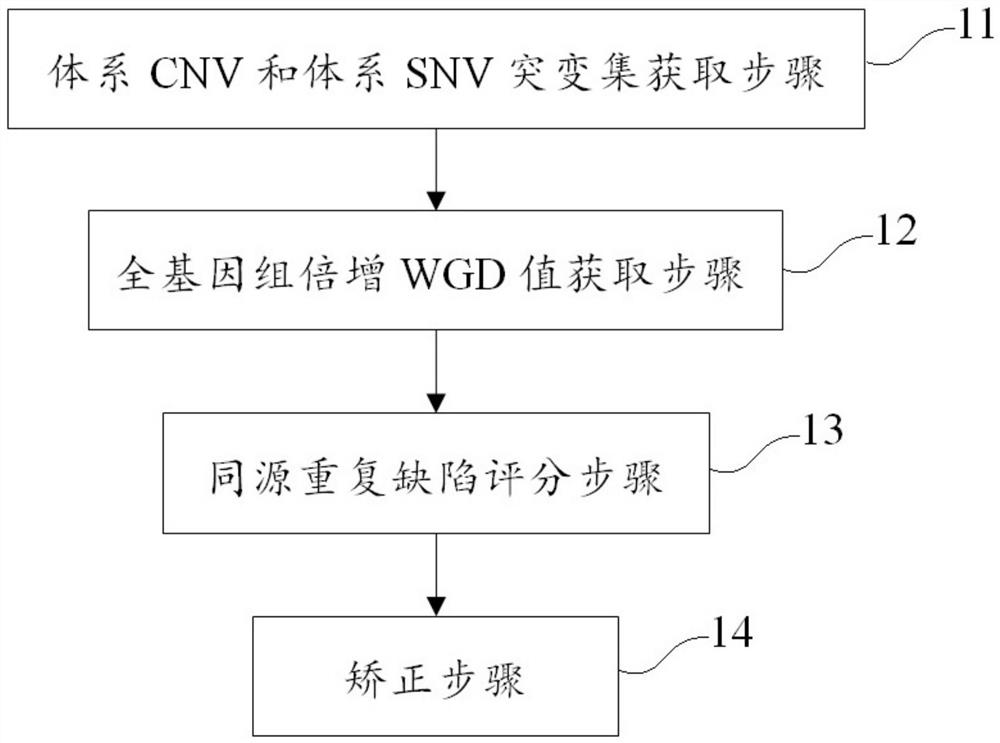
[0079] This example corrects the method of homologous recombination defect score, including the following steps:
[0080] System CNV and System SNV Mutant Setting Procedure: Get the system CNV mutation and system SNV mutation to be tested.
[0081] This example uses GATK software to detect CNV mutation, combined with the sample CNV result, filter embryo CNV mutation, to obtain a system CNV mutation; use MUTECT2 software to detect SNV mutation, combined with the sample SNV result, filter embryo SNV mutation, to obtain system SNV mutation .
[0082] All genome Pienza WGD value acquisition steps: including the use of the obtained system CNV mutation and system SNV mutation, calculate the WGD value under the optimal model of the sample to be tested.
[0083] This example uses the filtered system CNV, SNV mutation point set as an input source software ABSOLUTE input, outputs WGD values, purity values, PLOIDY values under multiple models to be tested, according to autonomous research ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com