Pharmacological prediction method for anti-tumor angiogenesis key targets of oldenlandia diffusa
A technology of Hedyotis diffusa and angiogenesis is applied in the field of pharmacological research of active ingredients of traditional Chinese medicine, which can solve the problems of lack of theoretical research, heavy workload, lack of pharmacological prediction methods, etc., and achieve the effect of clarifying the experimental direction and saving experimental resources.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
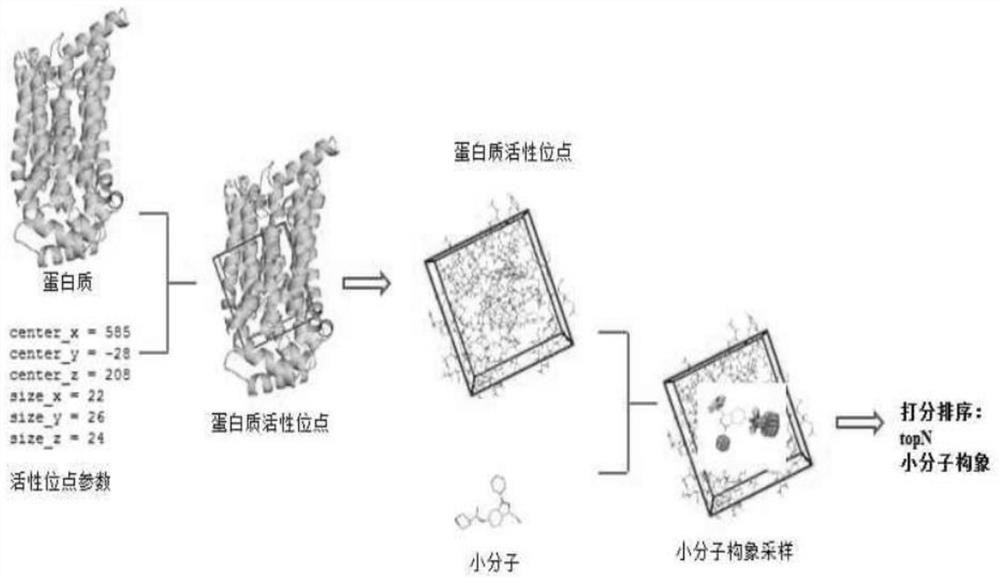
[0019] Pharmacological prediction method for key targets of anti-tumor angiogenesis of Hedyotis diffusa, including the following steps:
1) Building modules: the modules include the Hedyotis diffusa component database module and the anti-tumor angiogenesis target module;
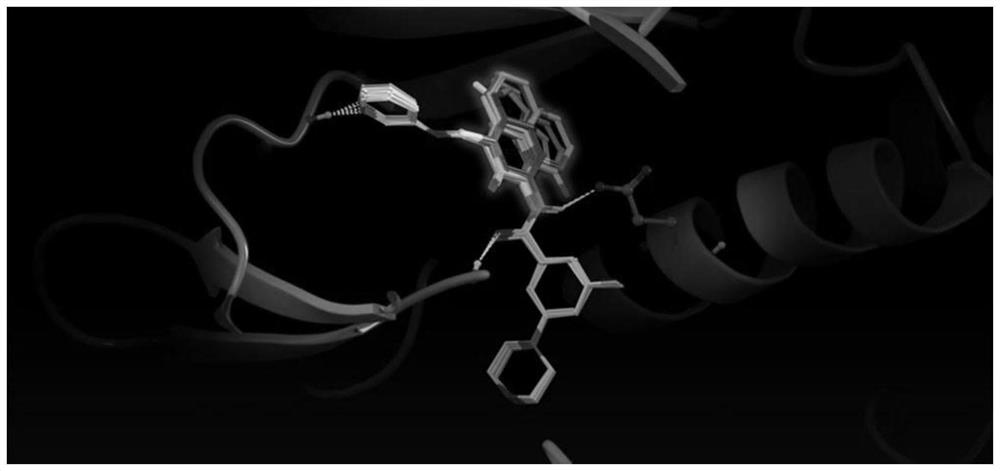
2) Using molecular docking software, batch read the components and targets in the module for molecular docking, and obtain the docking score; see the appendix for the general process. figure 1, Molecular docking is the process of mutual recognition between two or more molecules through geometric matching and energy matching. Molecular docking has shown very important application value in drug design. In the process of the interaction between enzyme activators, enzyme inhibitors and enzymes and the pharmacological reaction of drug molecules, small molecules (Ligand) and target (Receptor) are combined with each other. First, the two molecules need to be sufficiently close, and a suitable orientation should be ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com