Method for detecting presence of severe acute respiratory syndrome coronavirus 2 in biological sample
A technology for respiratory syndrome and biological samples, applied in the field of real-time identification of pathogenic microorganisms and/or their antibiotic resistance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
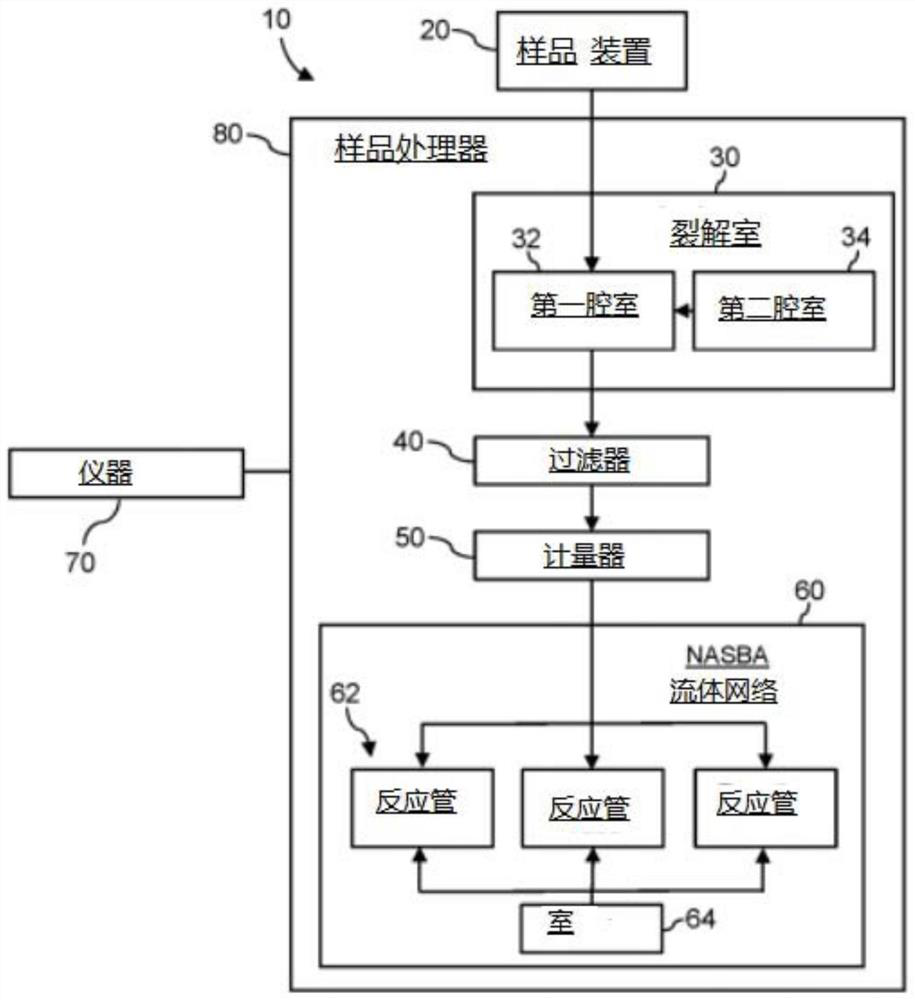
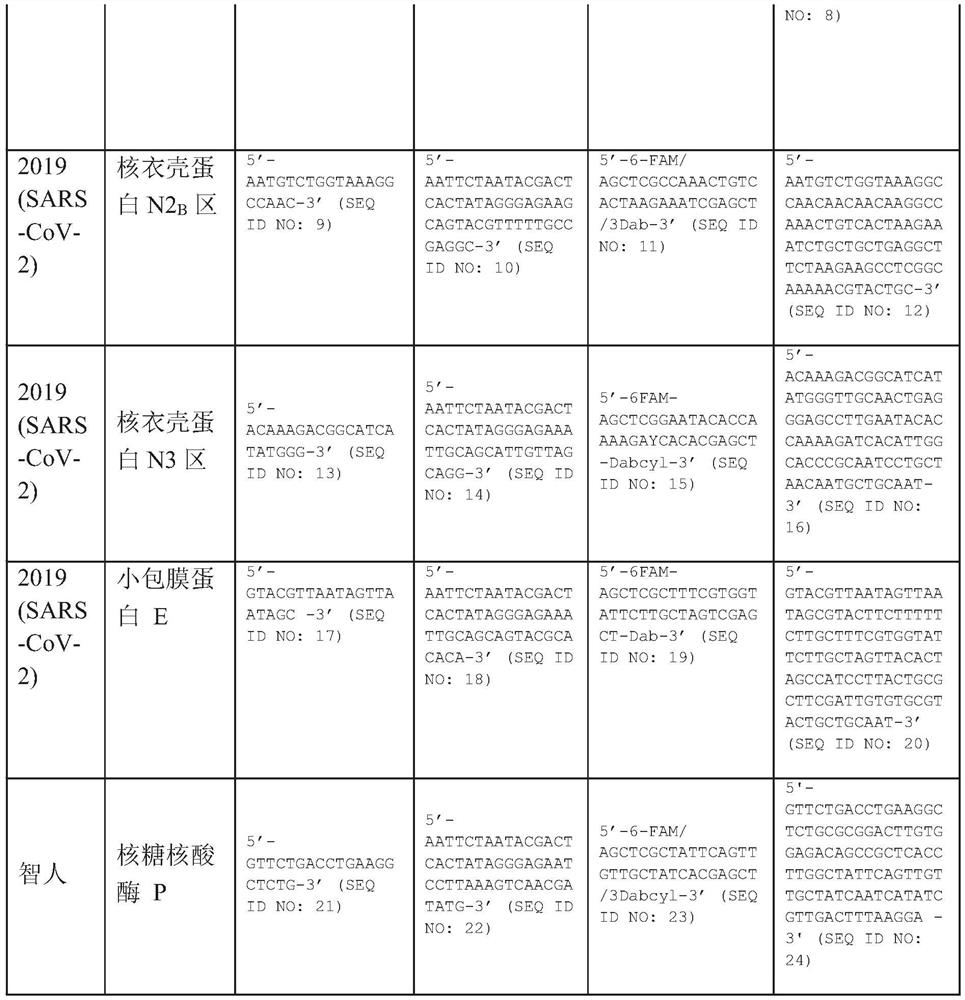
[0014] One aspect of this disclosure involves a method for detecting severe acute respiratory syndrome Coronatte 2 (SARS-COV-2) in biological samples (SARS-COV-2), which includes the following steps Cracks; the target nucleic acid sequences in the crack -based nucleic acid sequence (NASBA) are generated by existing the existence of positive primers, reverse primers and molecular beaches to generate expanded cracks. The positive primer has the oligonucleotide sequence of a group composed of a group of groups composed of a group composed of a group composed of a group. The reverse primer has the oligonucleotide sequence of the group of groups composed of a group. The molecular label has the oligonucleotide sequence and fluorescent group of the seq ID NO: 3, SEQ ID NO: 7, SEQID NO: 11, SEQ ID NO: 15, and SEQ ID NO: 19. The exposure of the exposure of the extension is exposed to the source of the excitement. Detects the fluorescence of fluorescent groups exposed to exposed to the exci...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com