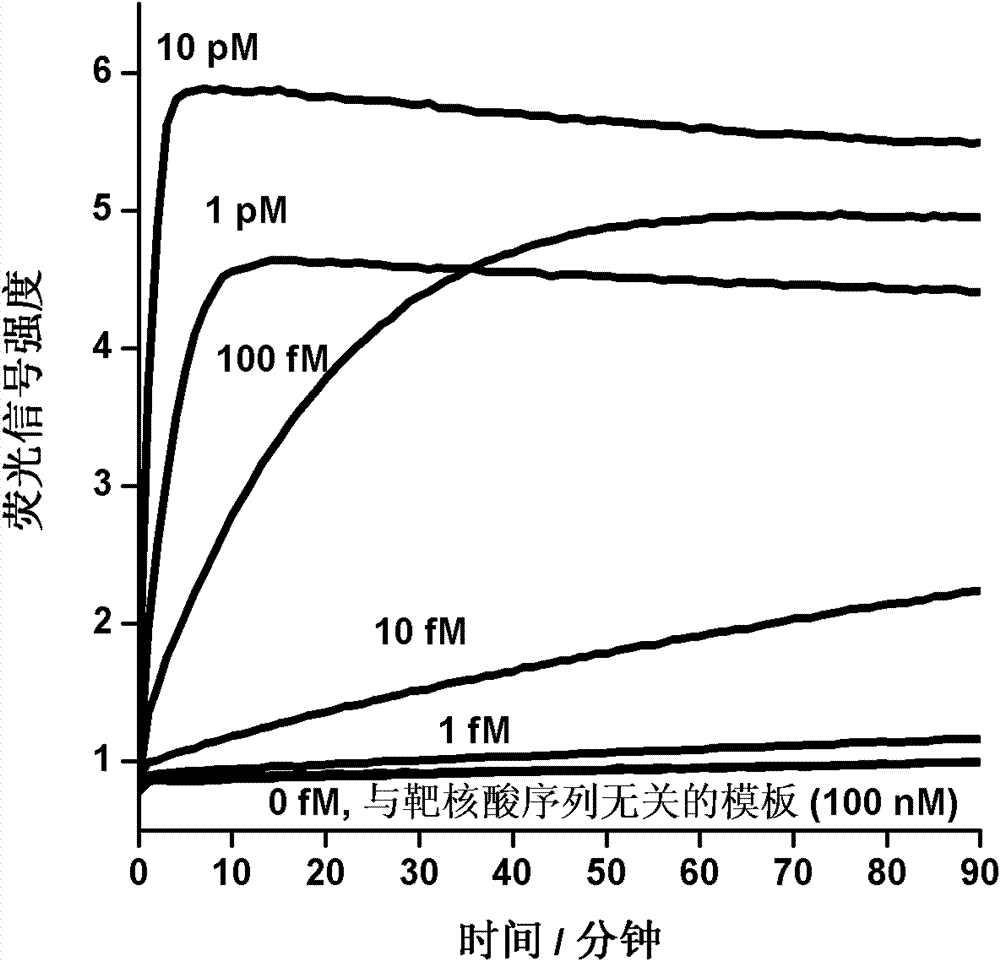
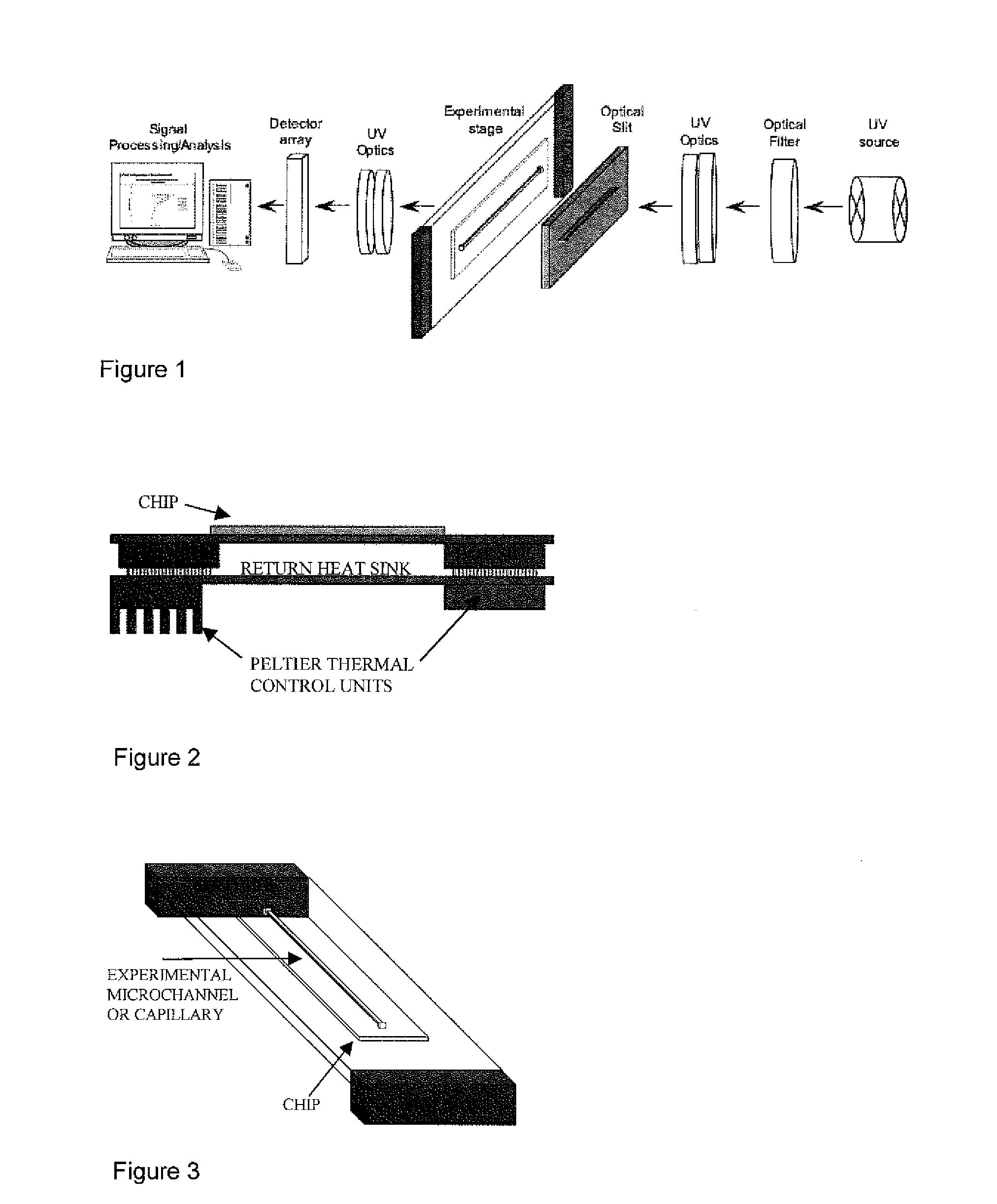
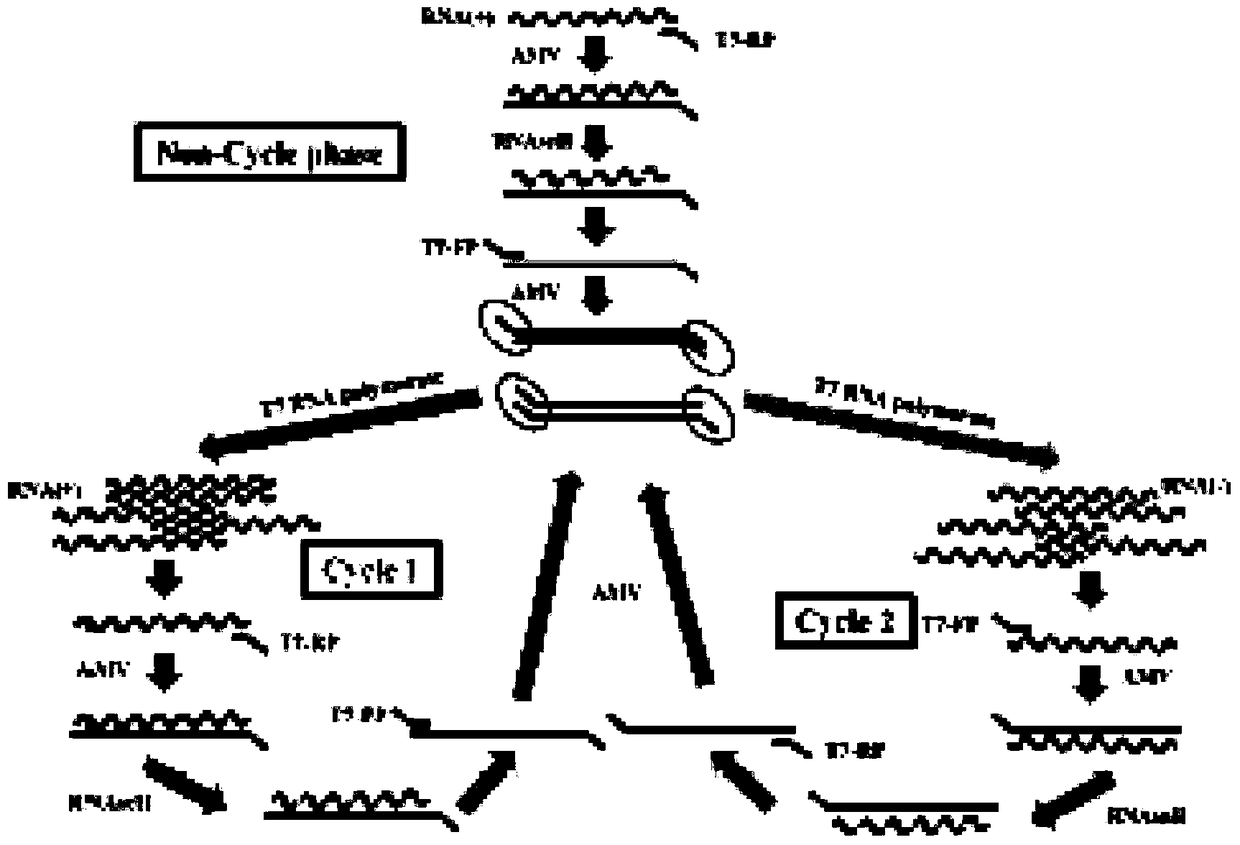
Patents
Literature
32 results about "Nucleic acid sequence based amplification" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Nucleic acid sequence based amplification (NASBA) is a method in molecular biology which is used to amplify RNA sequences. History. NASBA was developed by J Compton in 1991, who defined it as "a primer-dependent technology that can be used for the continuous amplification of nucleic acids in a single mixture at one temperature".
Method for amplifying nucleic acid sequence
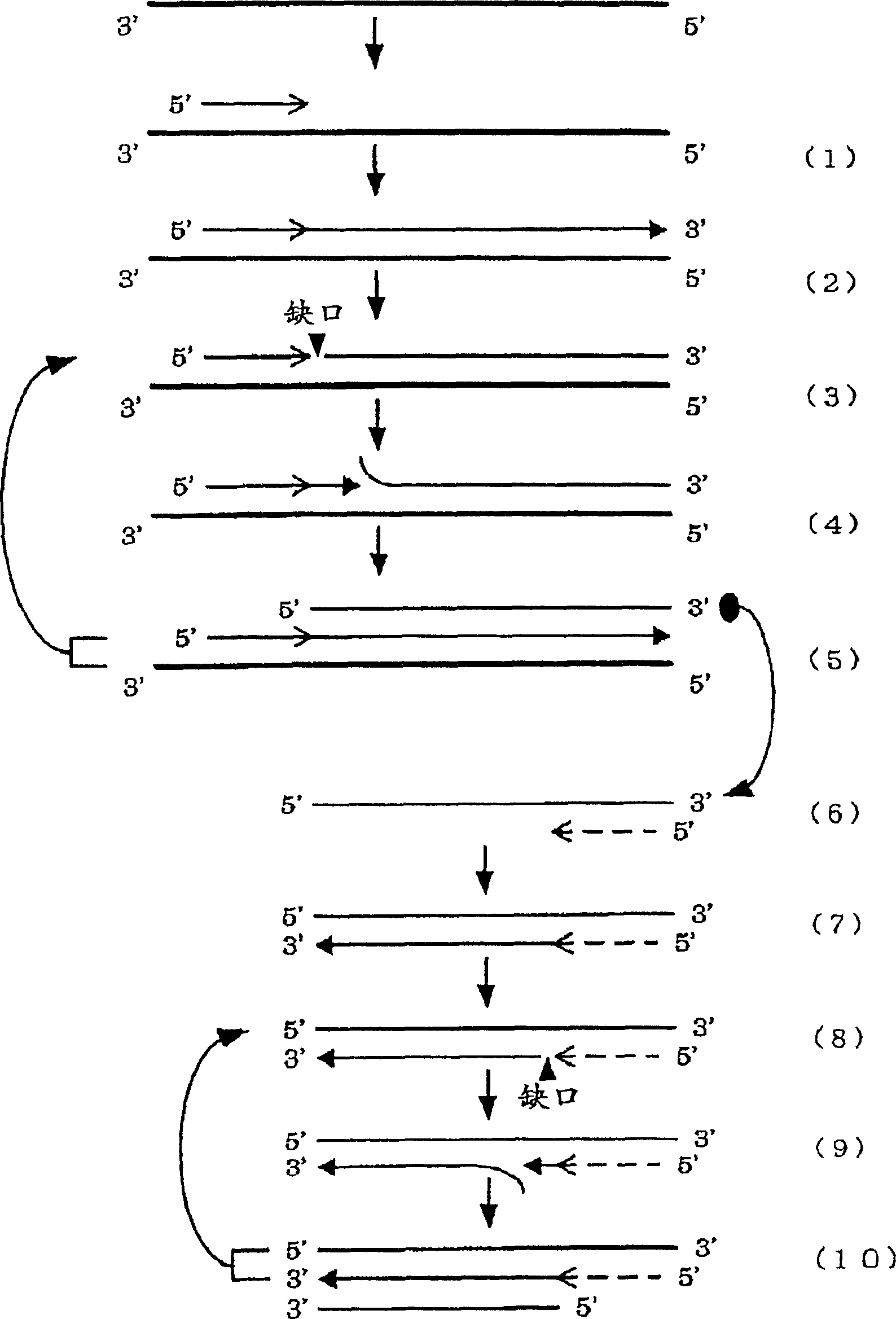
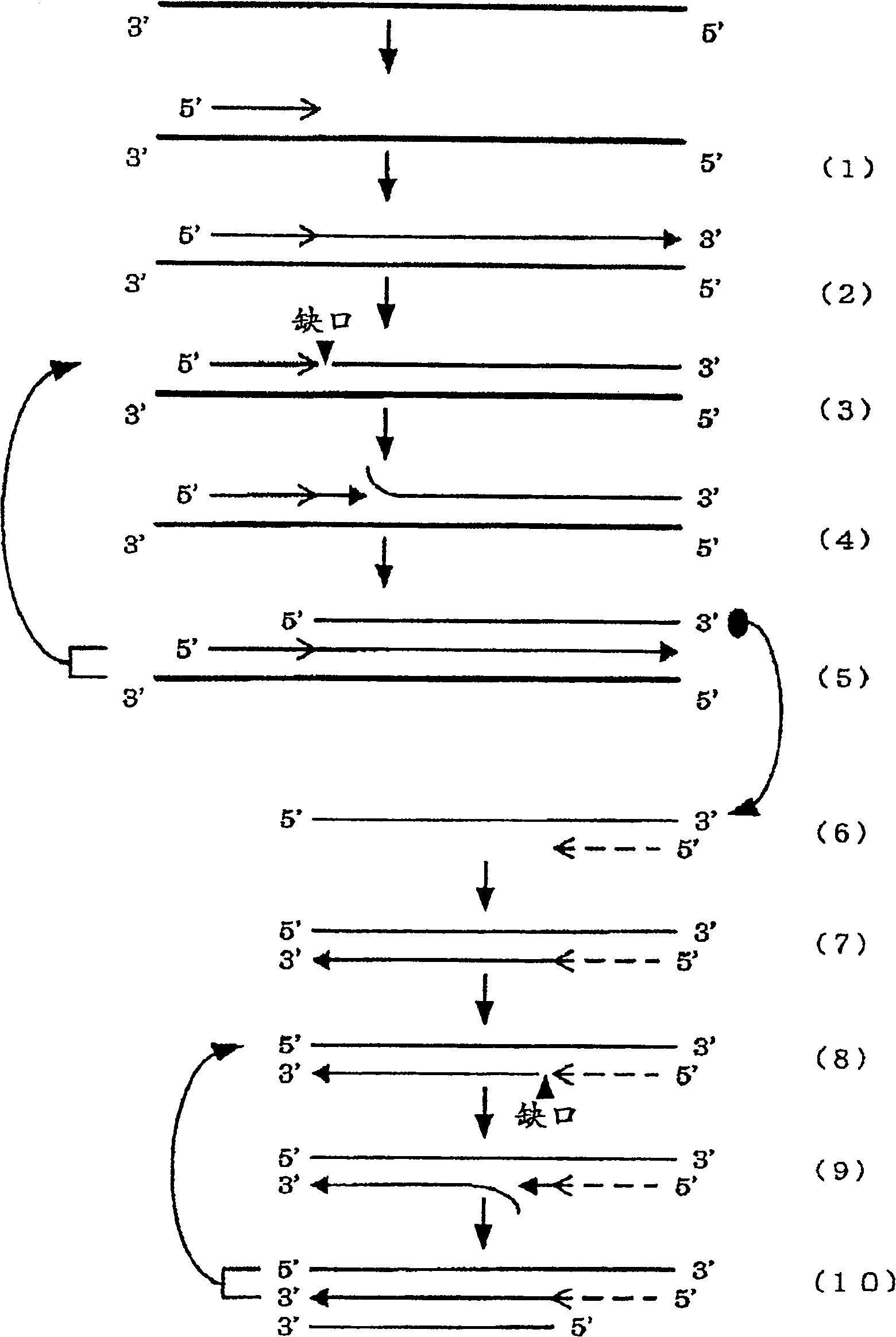
A convenient and effective method for amplifying a nucleic acid sequence characterized by effecting a DNA synthesis reaction in the presence of chimeric oligonucleotide primers; a method for supplying a large amount of DNA amplification fragments; an effective method for amplifying a nucleic acid sequence by combining the above method with another nucleic acid sequence amplification method; a method for detecting a nucleic acid sequence for detecting or quantitating a microorganism such as a virus, a bacterium, a fungus or a yeast; and a method for detecting a DNA amplification fragment obtained by the above method in situ.
Owner:TAKARA HOLDINGS
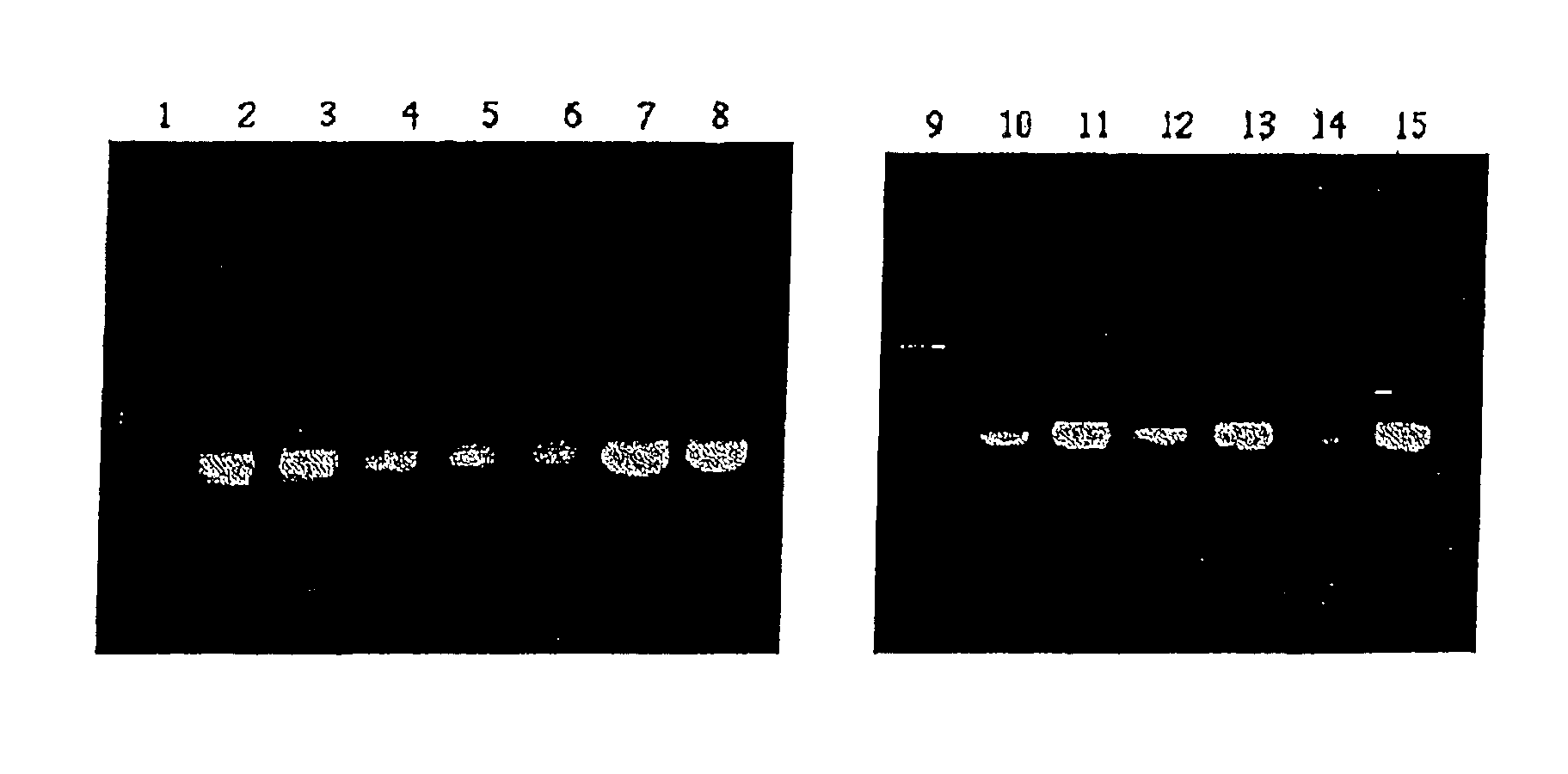
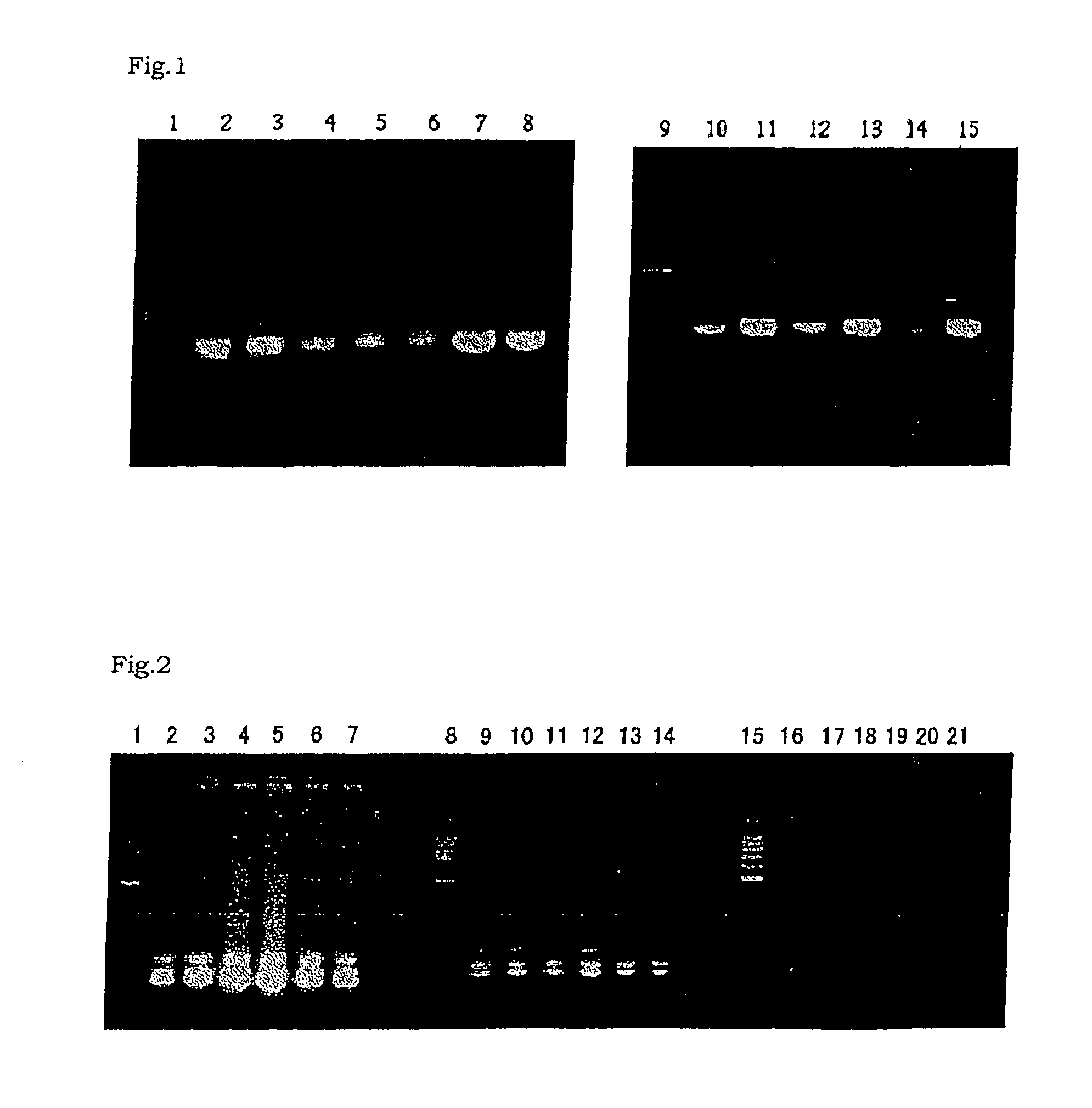
Method and kit for detecting pathogenic bacterium of food source by test strip based on NASBA (Nucleic Acid Sequence Based Amplification)
ActiveCN103146835AImprove featuresHigh sensitivityMicrobiological testing/measurementMicroorganism based processesNucleic acid sequence based amplificationMicrobiology
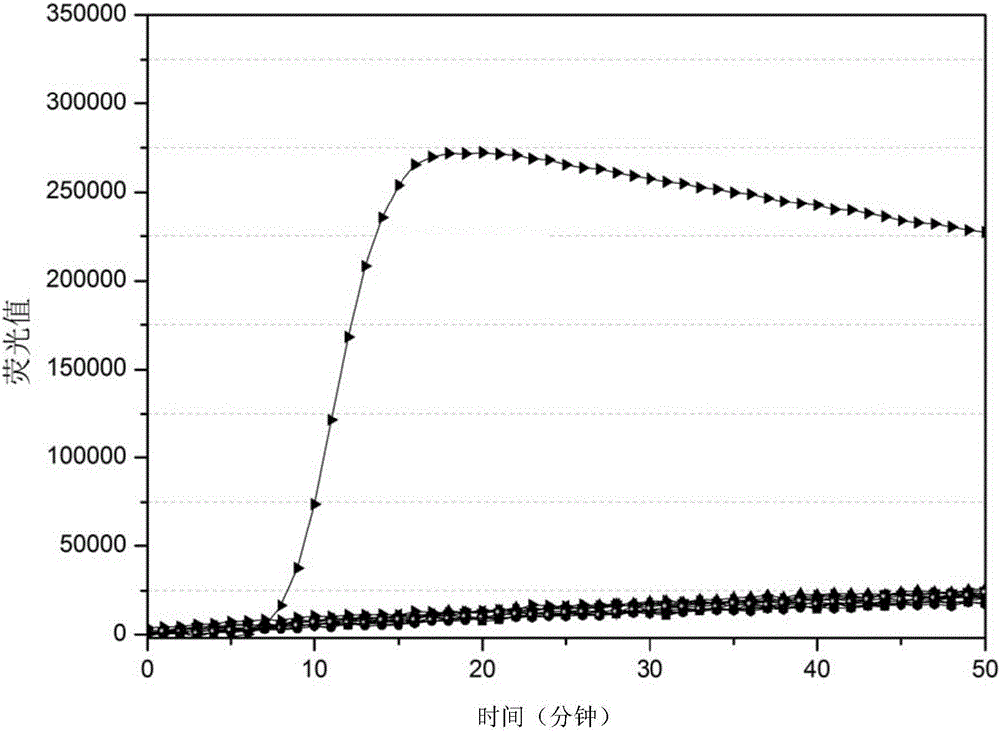
The invention discloses a method and a kit for detecting pathogenic bacterium of a food source by a test strip based on NASBA (Nucleic Acid Sequence Based Amplification) and belongs to the technical field of biological detection. The method comprises the following steps of: (1) extracting RNA of pathogenic bacterium of the food source; (2) designing an amplification primer; (3) performing NASBA reaction; (4) designing a catching probe; (5) preparing a nano-gold probe; (6) preparing a collaurum nucleic acid strip; and (7) detecting the samples. The kit using the method to detect Listeria monocytogenes comprises primers (as shown in SEQ ID NO. 1 and 2), enzyme, buffer solution, RNaseH, RNA enzyme inhibitor, dNTPs, NTPs, probes (as shown in SEQ ID NO. 4, 5 and 6) and collaurum nucleic acid strip, etc. By combining NASBA with detection of the test strip, the method disclosed by the invention can realize qualitative or quantitative detection, and has low cost, high detection speed, good specificity, high sensitivity and safety in use.
Owner:SOUTH CHINA NORMAL UNIVERSITY
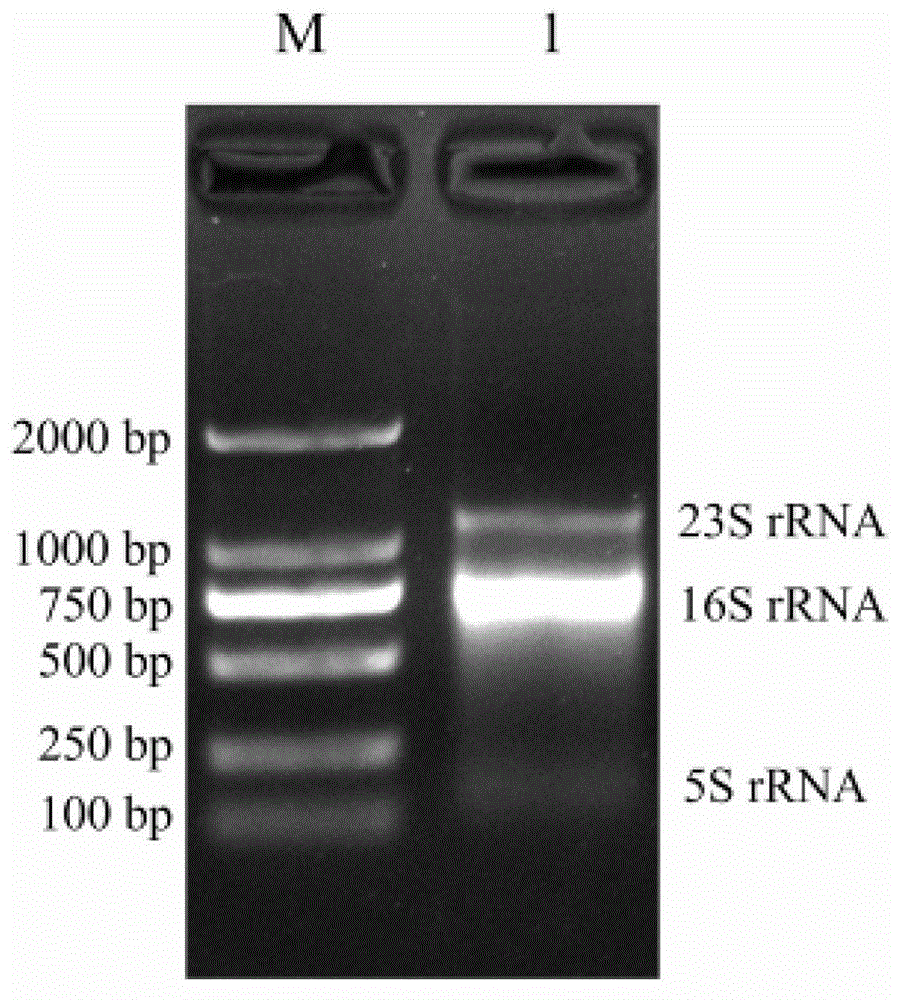
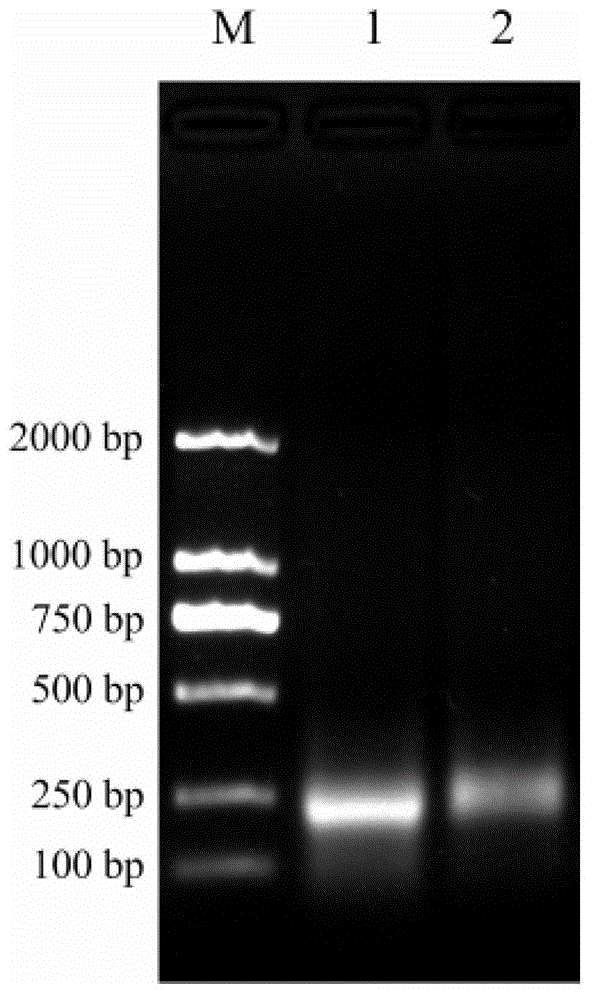
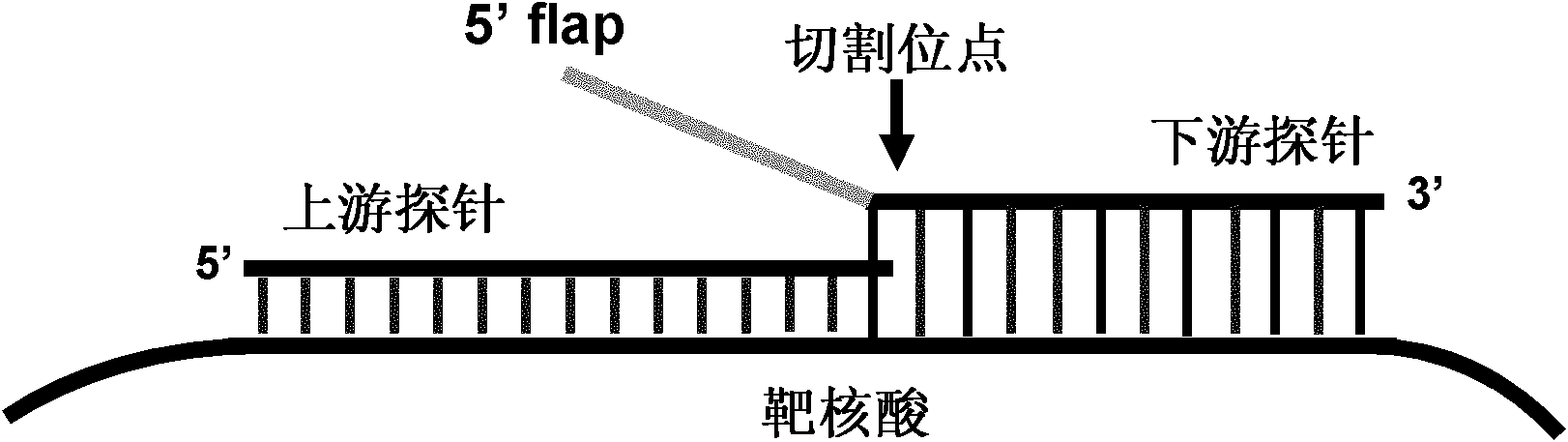
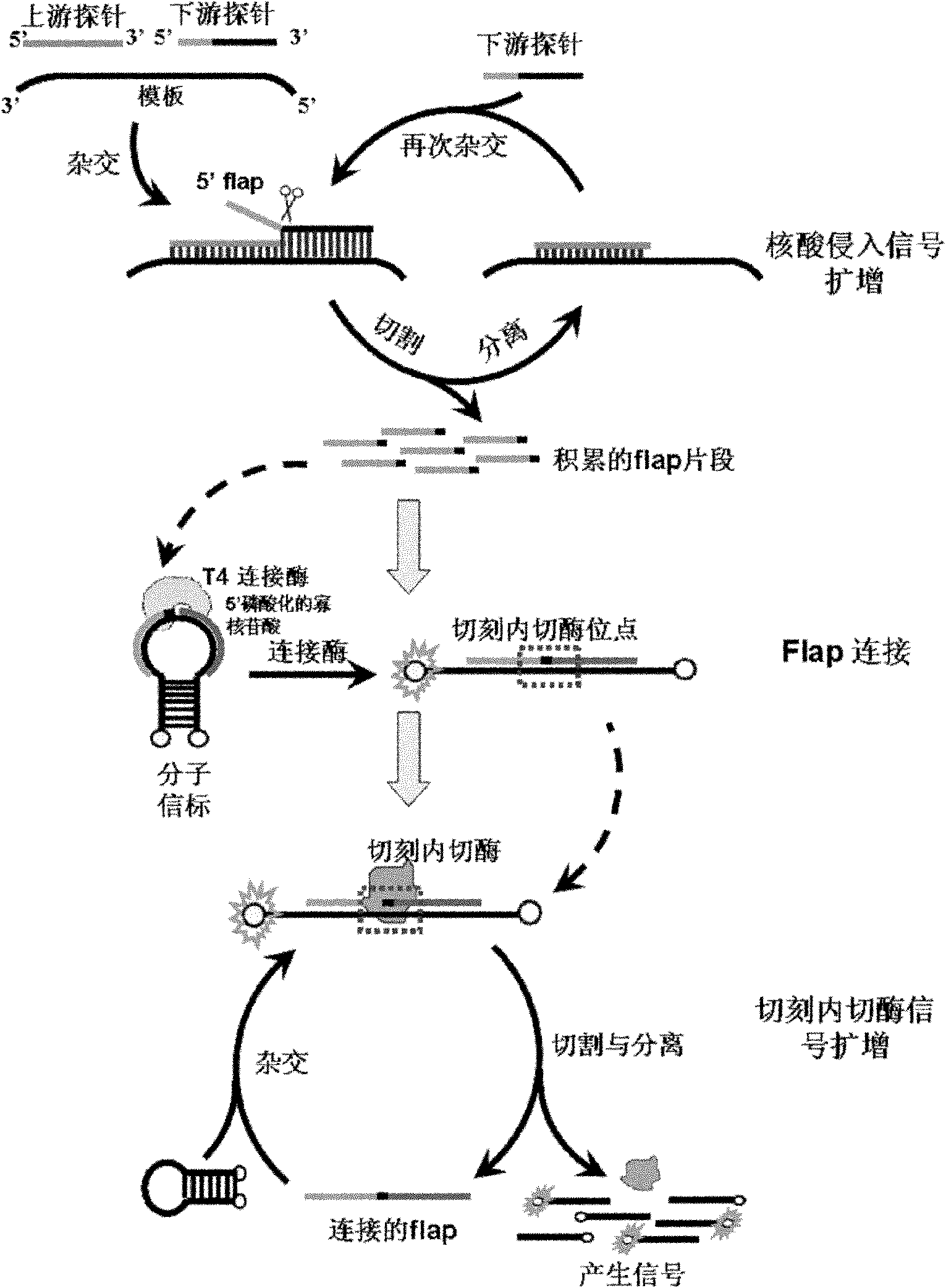
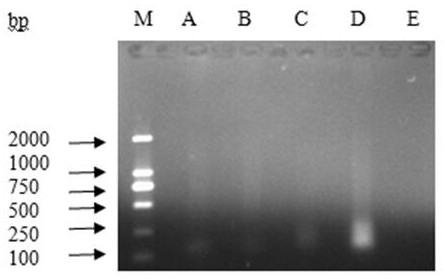
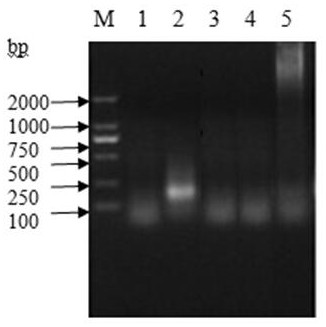
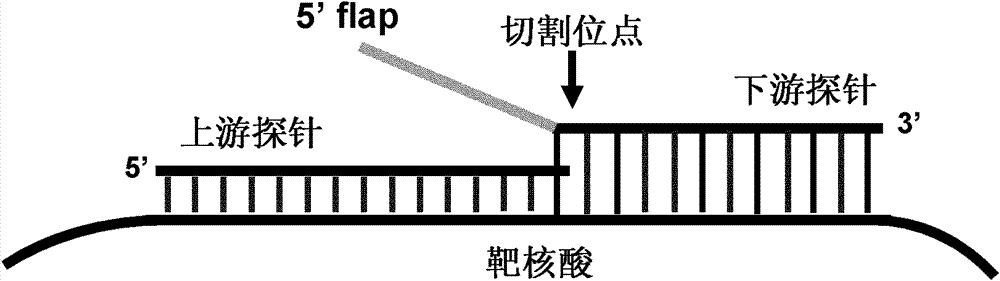
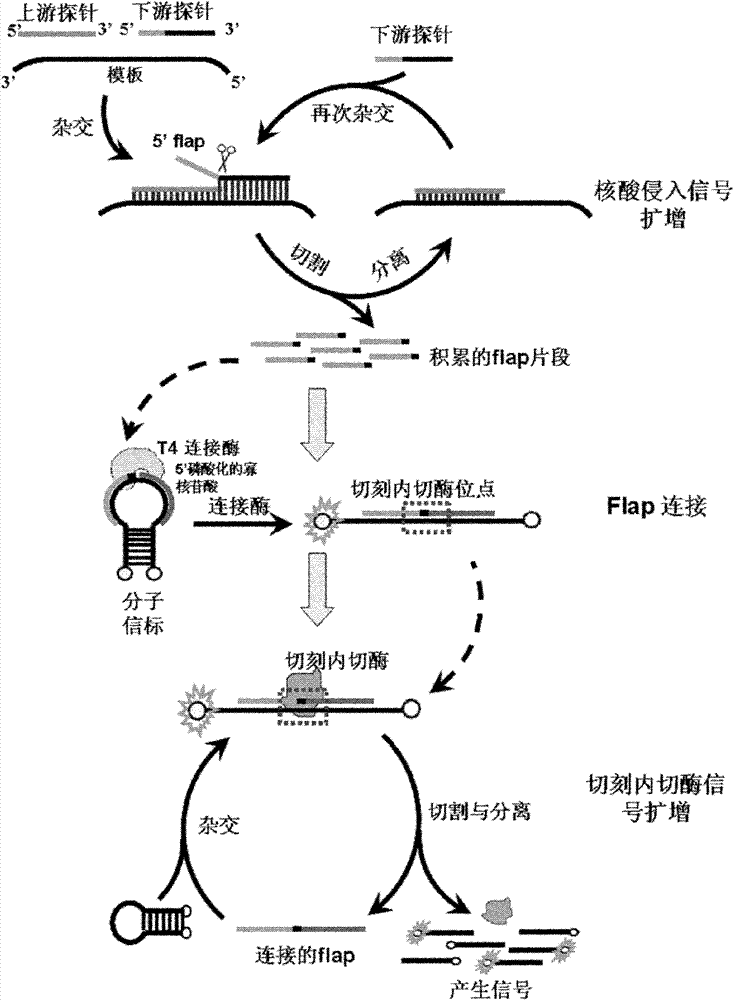
Method for inspecting nucleic acid signal amplification of ligation nucleic acid intrusive reaction and cutting endonuclease reaction
InactiveCN101974638ALow costNo cross contaminationMicrobiological testing/measurementFluorescence/phosphorescenceFluorescenceGenetics
The invention discloses a method for detecting nucleic acid signal amplification of ligation nucleic acid intrusive reaction and cutting endonuclease reaction, which belongs to the field of molecular biology. The method comprises the following steps: 1) during nucleic acid intrusive signal amplification, accumulating flap fragments; 2) during flap fragment connection, connecting the flap fragments with another oligonucleotide on a molecular beacon to form a cutting endonuclease site; 3) during cutting endonuclease signal amplification, only cutting a molecular beacon chain so that the fluorescent group of the molecular beacon is separated from a quenching group, and a new integrated molecular beacon is hybridized with a flap fragment connected with the molecular beacon, thus amplifying a target nucleic acid signal; and 4) inspecting the target nucleic acid signals. The method is capable of inspecting nucleic acid target sequences and respectively inspecting nucleic acid sequences with single base difference near intrusive sites.
Owner:HUADONG RES INST FOR MEDICINE & BIOTECHNICS
NASBA (Nucleic Acid Sequence Based Amplification) detection primer, probe and detection method for GII type norovirus in sea foods
PendingCN103525947ANo cross reactionEasy to operateMicrobiological testing/measurementMicroorganism based processesForward primerNucleic acid sequence based amplification
The invention discloses an NASBA (Nucleic Acid Sequence Based Amplification) detection primer, a probe and a detection method for GII type norovirus in sea foods. According to the NASBA detection primer, a forward primer sequence is represented as SEQ ID NO.1 and a backward primer sequence is represented as SEQ ID NO.2; the primer sequence of a molecular beacon probe for detecting the GII type norovirus is represented as SEQ ID NO.3. The detection method comprises the following steps: (1) constructing a standard cRNA (complementary Ribonucleic Acid) of the GII type norovirus; (2) carrying out a real-time NASBA reaction; (3) qualitatively and quantitatively analyzing. The primer and the probe disclosed by the invention have high specificity on the detection of the GII type norovirus. The detection method is simple in operation steps and short in detection time, can be used for carrying out a qualitative and quantitative analysis and has high specificity and sensitivity.
Owner:YELLOW SEA FISHERIES RES INST CHINESE ACAD OF FISHERIES SCI
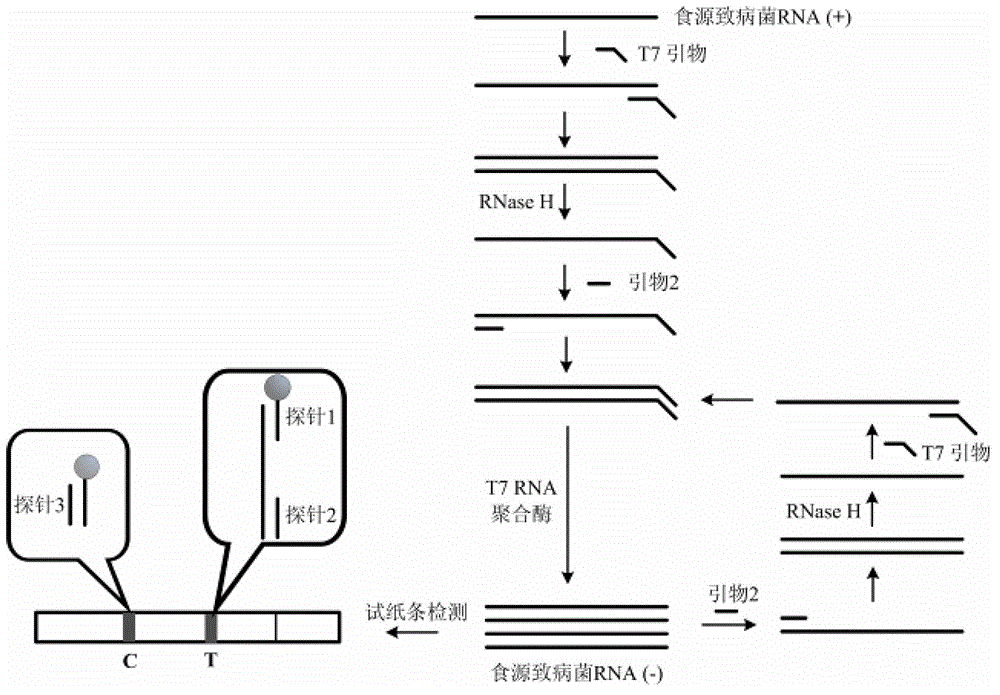
Novel high-sensitivity respiratory virus nucleic acid NASBA (nucleic acid sequence based amplification) primers and detection method
ActiveCN105256068AHigh detection sensitivityEliminate hazardsMicrobiological testing/measurementDNA/RNA fragmentationThroat swab sampleT7 RNA polymerase
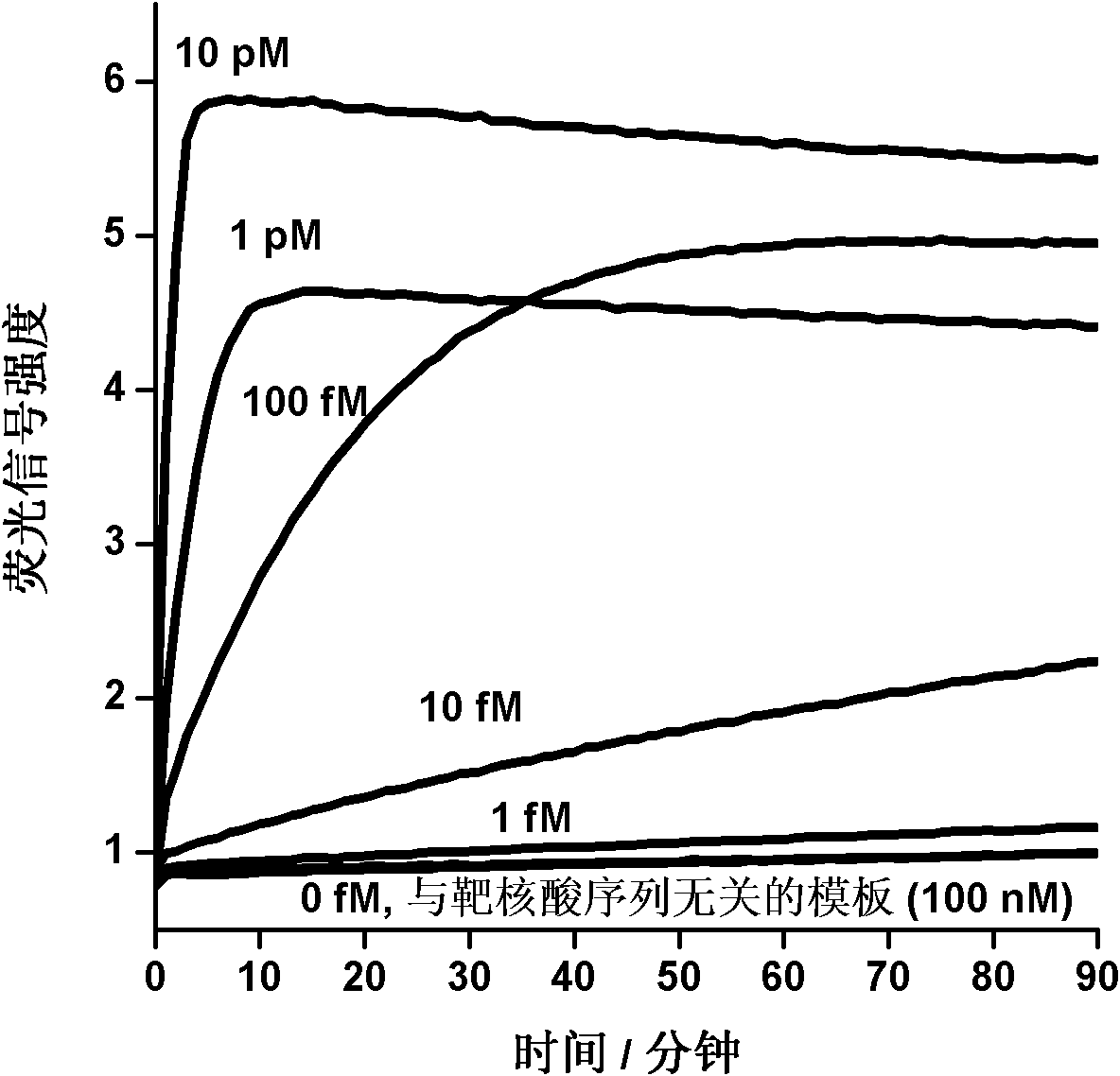
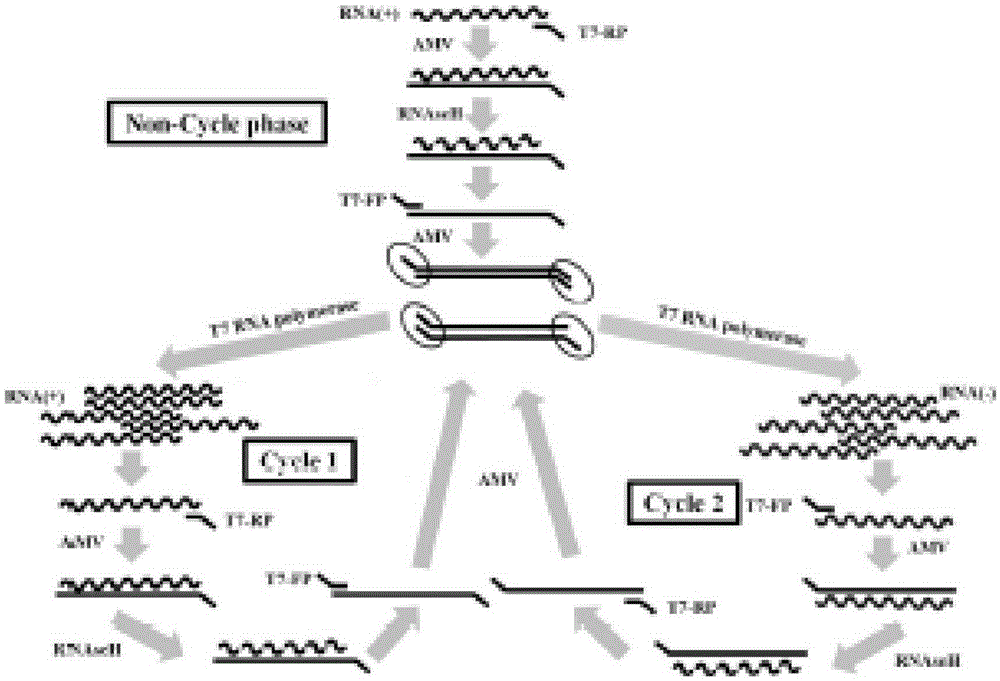
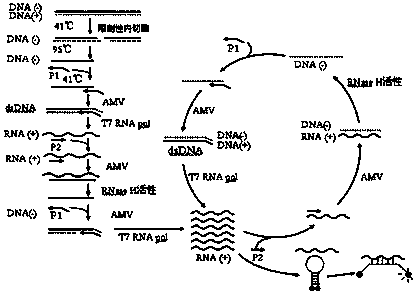
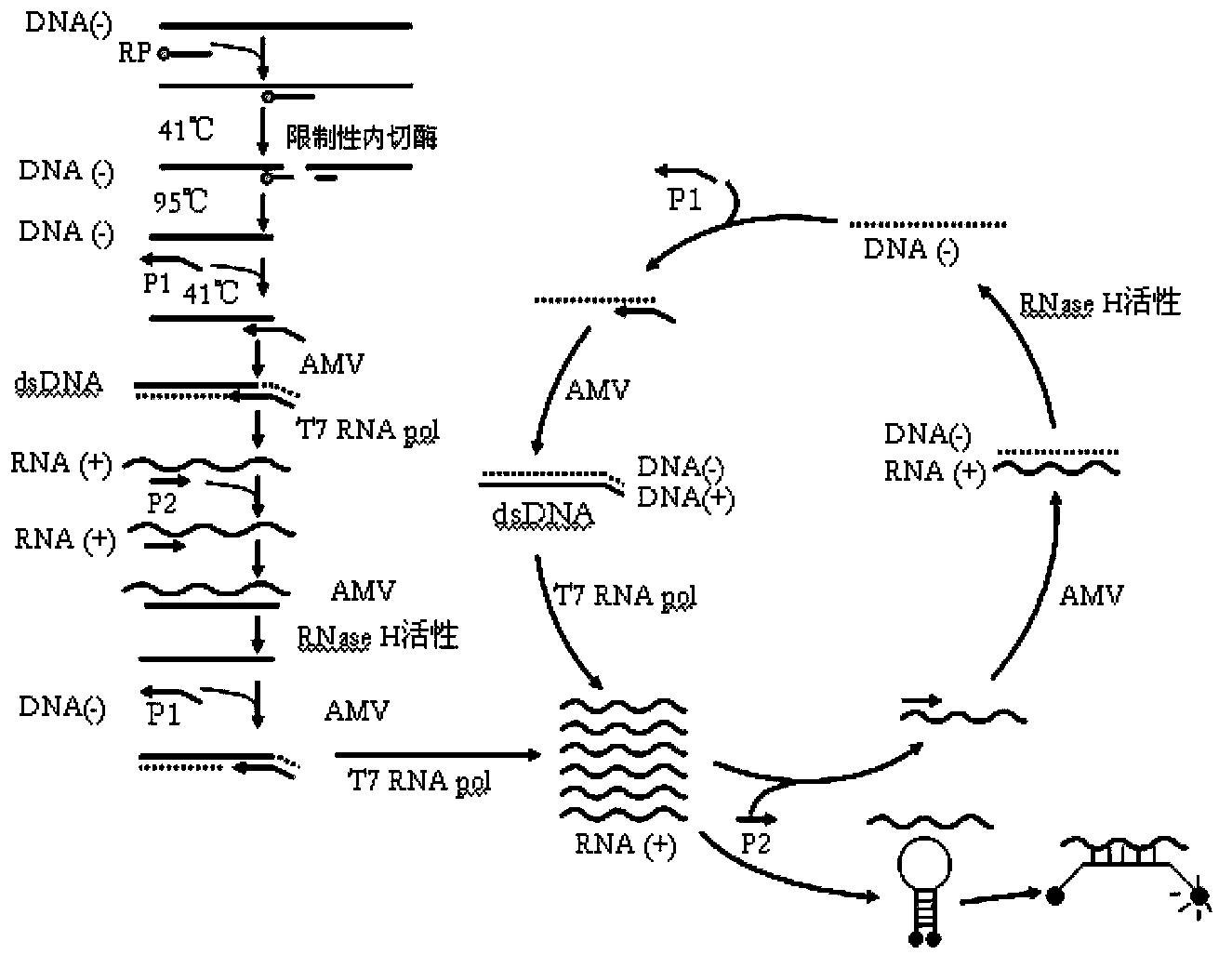
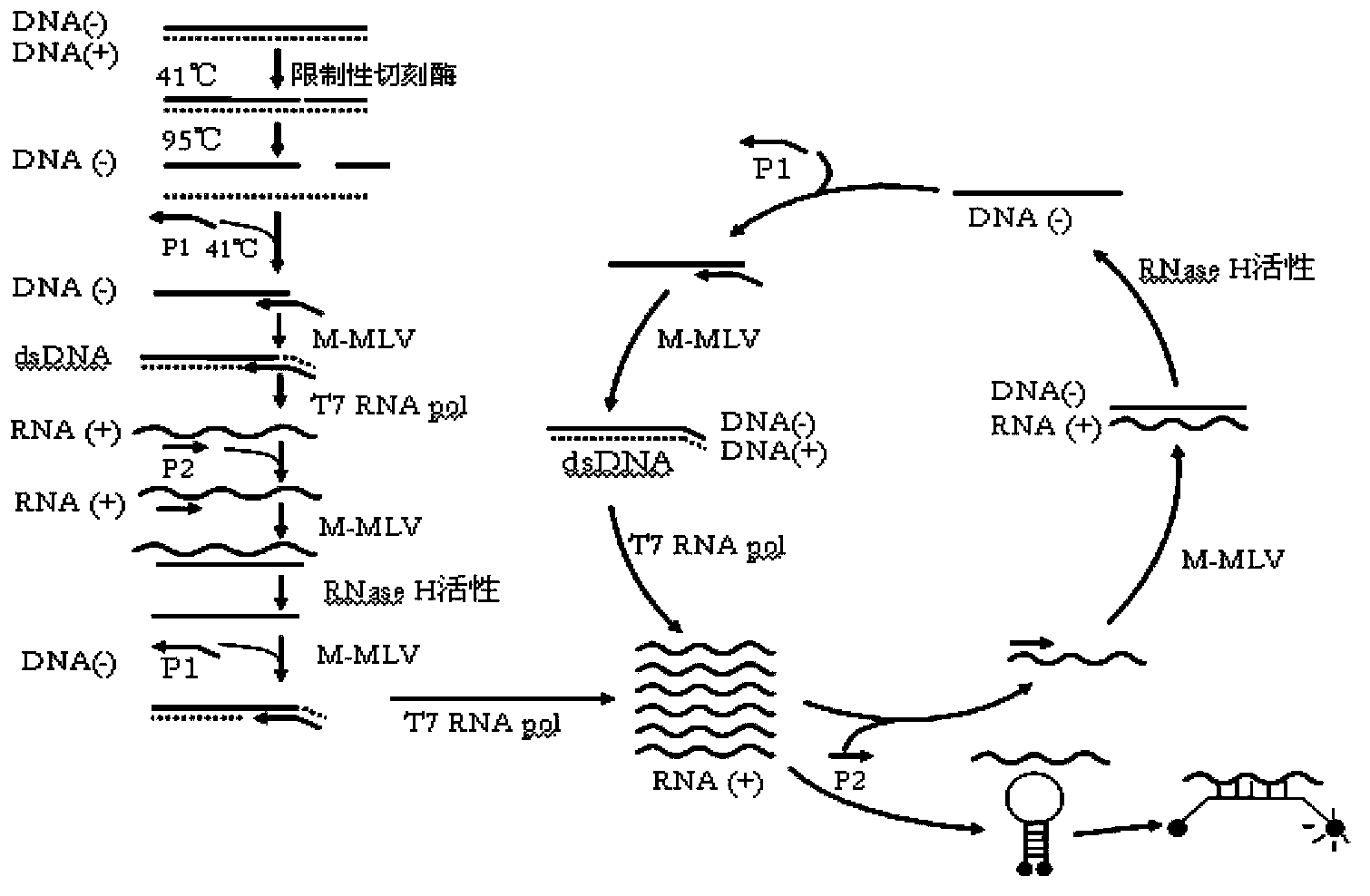
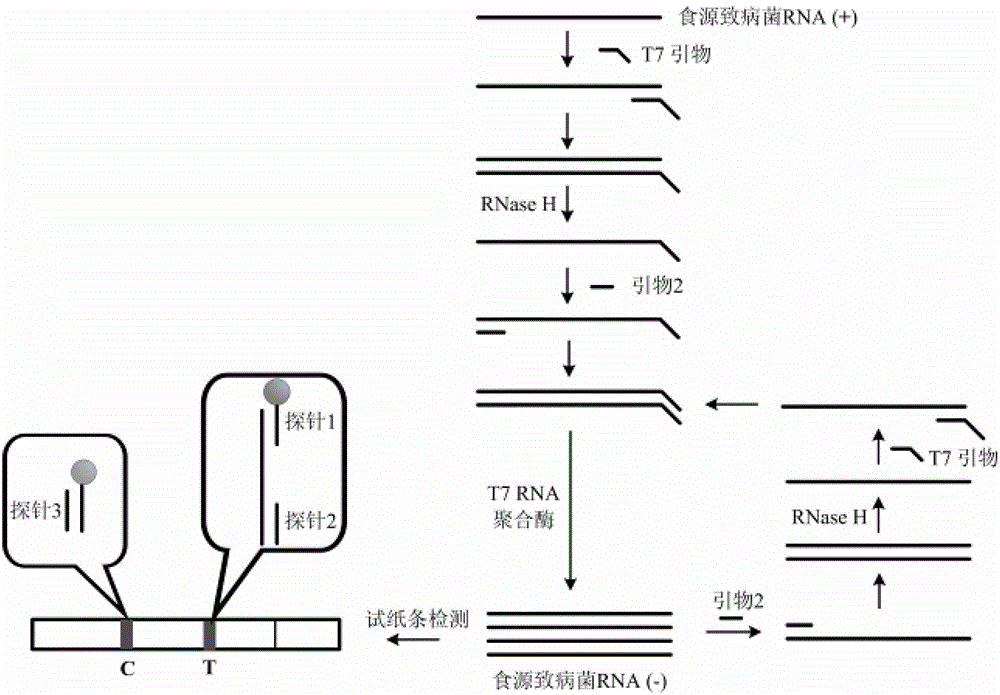
The invention provides novel high-sensitivity respiratory virus nucleic acid NASBA (nucleic acid sequence based amplification) primers. A respiratory virus conservative area is selected, and a promoter sequence capable of being recognized by T7 RNA (ribose nucleic acid) polymerase is formed at the 5' terminal of one of the synthesized primers. The invention further provides a novel high-sensitivity respiratory virus nucleic acid NASBA detection method. T7 RNA polymerase promoter sequences are added to the 5' terminals of the two primers complementary to two ends of a respiratory virus RNA sequence, so that when the primers enter a circulation phase, RNA sequences [RNA(+)] consistent with a template sequence can be synthesized, RNA sequences [RNA(-)] complementary to a template RNA sequence can be synthesized, and the detection sensitivity of viral nucleic acid is greatly improved; a throat swab sample of a patient can be directly used for amplification, a reverse transcription process is not required, and the operation time is saved.
Owner:HANGZHOU JOINSTAR BIOTECH
Hand-foot-mouth disease virus EV71 nucleic acid detection kit
InactiveCN102676692AHigh sensitivityNo pollutionMicrobiological testing/measurementMicroorganism based processesReverse transcriptaseTotal rna
The invention discloses a hand-foot-mouth disease virus EV71 nucleic acid detection kit, and belongs to the technical field of virus detection. The kit consists of N.BstNBI enzyme, T7 RNA (Ribonucleic Acid) polymerase, AMV (Avian Myeloblastosis Virus) reverse transcriptase, RNase H, NASBA (Nucleic Acid Sequence Based Amplification) reaction solution, a NASBA reaction primer, an RIDA (Radioisotope Dilution Assay) probe, DEPC (Diethypyrocarbonate) water, influenza virus a HINI RNA, breast cancer cell MCF-7 total RNA and EV71 RNA. According to the kit, a hand-foot-mouth disease virus EV71 nucleic acid sequence is detected by combining NASBA and RIDA technologies; and the kit has the advantages of sensitivity, specificity, easiness, convenience, high speed, high efficiency, no dependency on special instruments and the like.
Owner:INST OF BASIC MEDICAL SCI ACAD OF MILITARY MEDICAL SCI OF PLA
Detection method and a kit of transgenic soybean MON89788 transformation event
InactiveCN103409498AReduce testing costsReduce mutual contaminationMicrobiological testing/measurementNucleic acid sequence based amplificationGenetically modified soybean
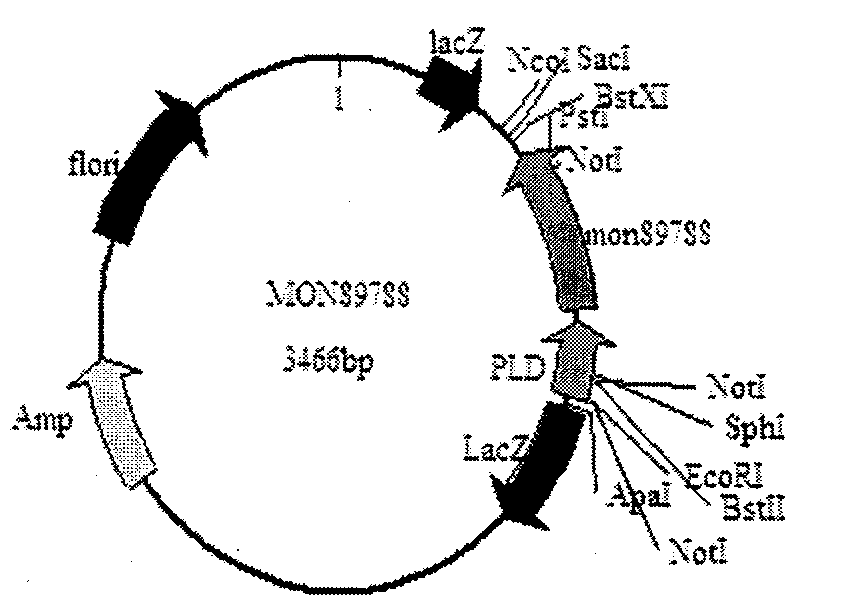
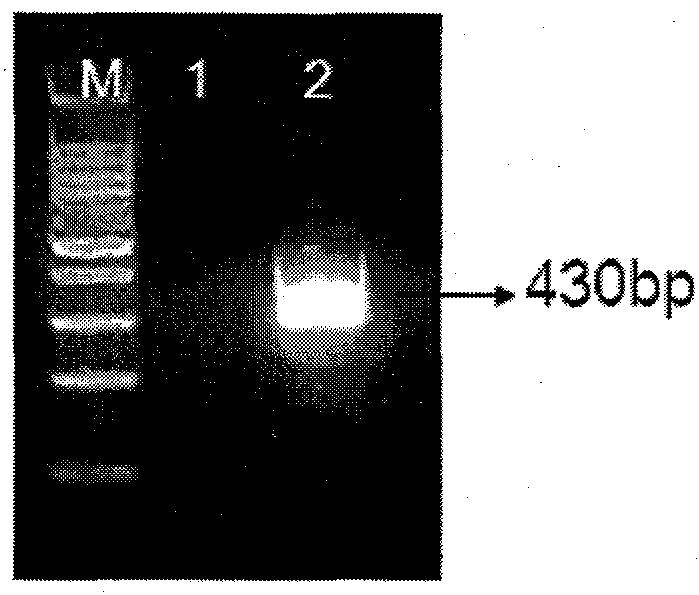
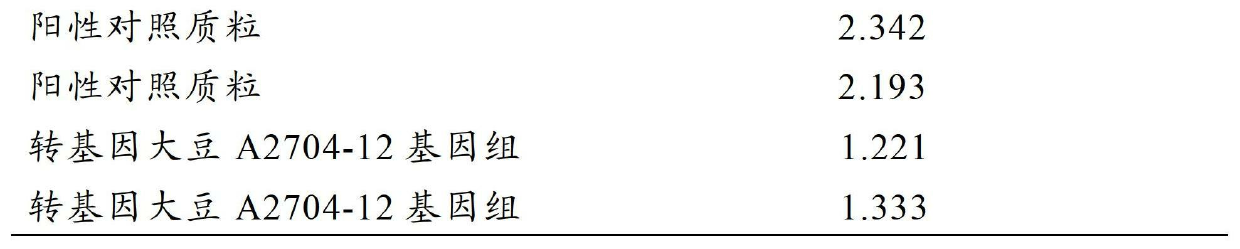
The invention discloses a detection method and a kit of transgenic soybean MON89788 transformation event. The method comprises the following steps: performing high temperature degeneration on a sample DNA (deoxyribonucleic acid), transcribing under RNA (ribonucleic acid) polymerase by using a primer for identifying MON89788 specificity region to generate RNA, performing NASBA (nucleic acid sequence based amplification) by using the above primer based on obtained RNA as a template; and finally detecting the amplification product. Without purchasing large instrument, the detection cost of the method disclosed by the invention is reduced, the detected product is easy to degrade, and the problem of cross contamination among samples is reduced.
Owner:海康生物科技(北京)有限公司 +1
Genotype determination method
InactiveCN102549171ASafety judgmentQuick and cheap judgmentMicrobiological testing/measurementRecombinant DNA-technologyReverse transcriptasePolymerase L
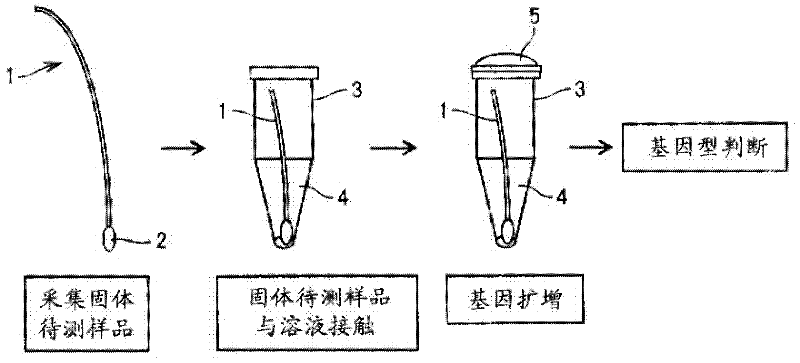
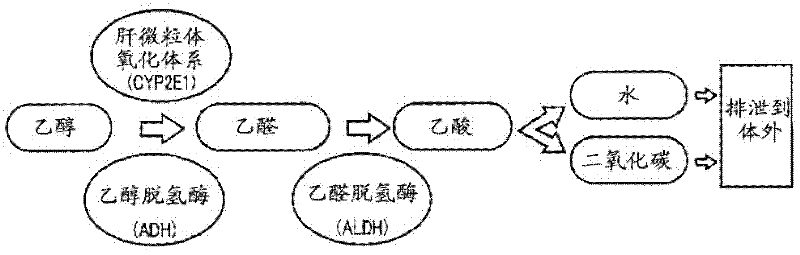
Disclosed is a method for determining a genotype of a gene associated with alcohol susceptibility, which comprises the steps of: bringing a solid test sample containing the gene into direct contact with a solution comprising a buffer and a DNA polymerase and primer DNA for the amplification of the gene to carry out a method selected from a polymerase chain reaction, an LAMP method, a chain substitution amplification method, a reverse transcriptase chain substitution amplification method, a reverse transcriptase polymerase chain reaction, a reverse transcription LAMP method, an amplification method based on a nucleotide sequence, a transcription-mediated amplification method, and a rolling circle-type amplification method, thereby amplifying the gene; and determining the genotype of the gene using the amplified gene. The method enables the determination of the genotype of a gene associated with alcohol susceptibility safely, rapidly and at low cost.
Owner:EDUCATIONAL CORP MUKOGAWA GAKUIN +1
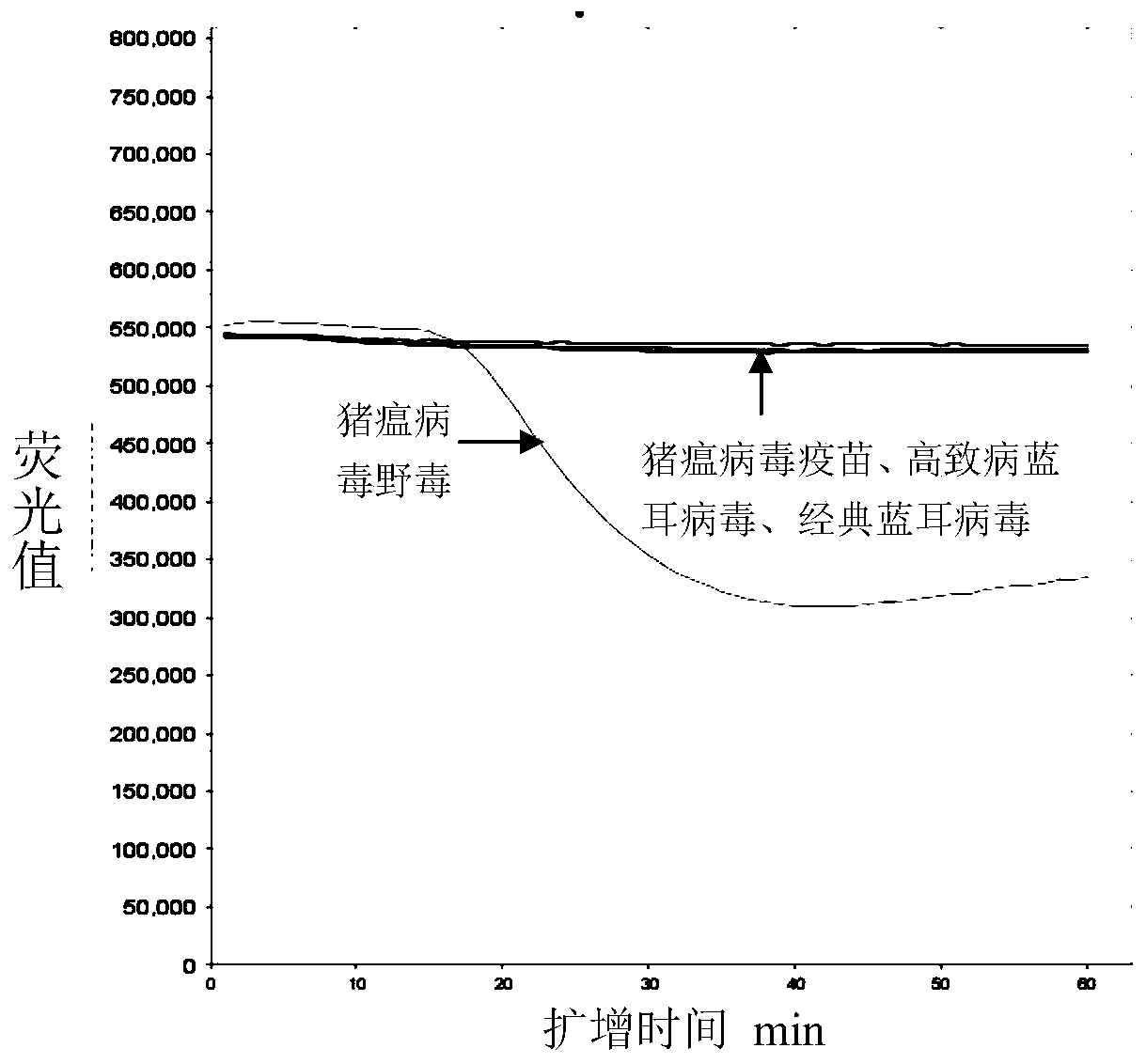
Kit for detecting wild strain of classical swine fever virus
ActiveCN110527747AMicrobiological testing/measurementAgainst vector-borne diseasesTreatment feverNucleic acid sequence based amplification
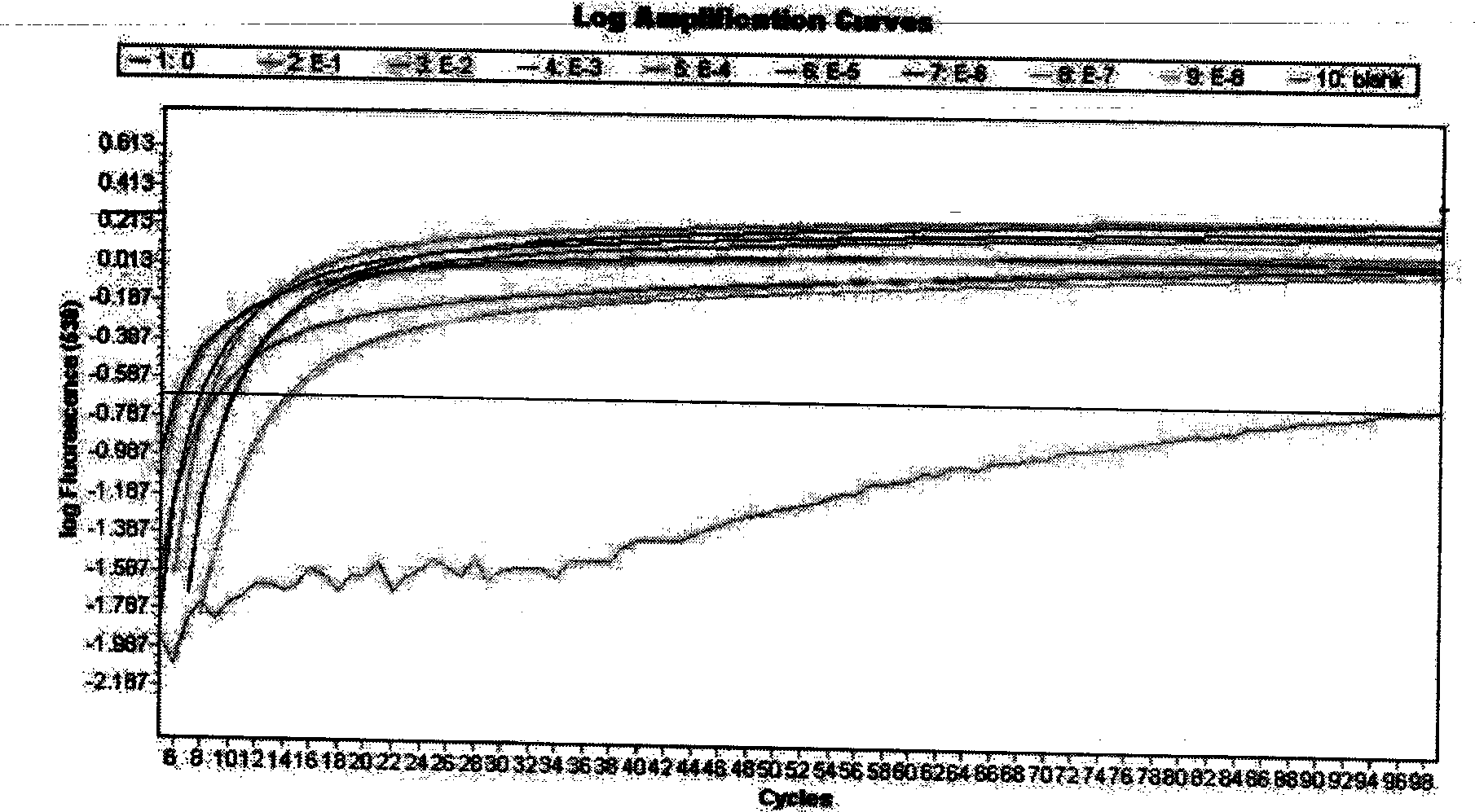
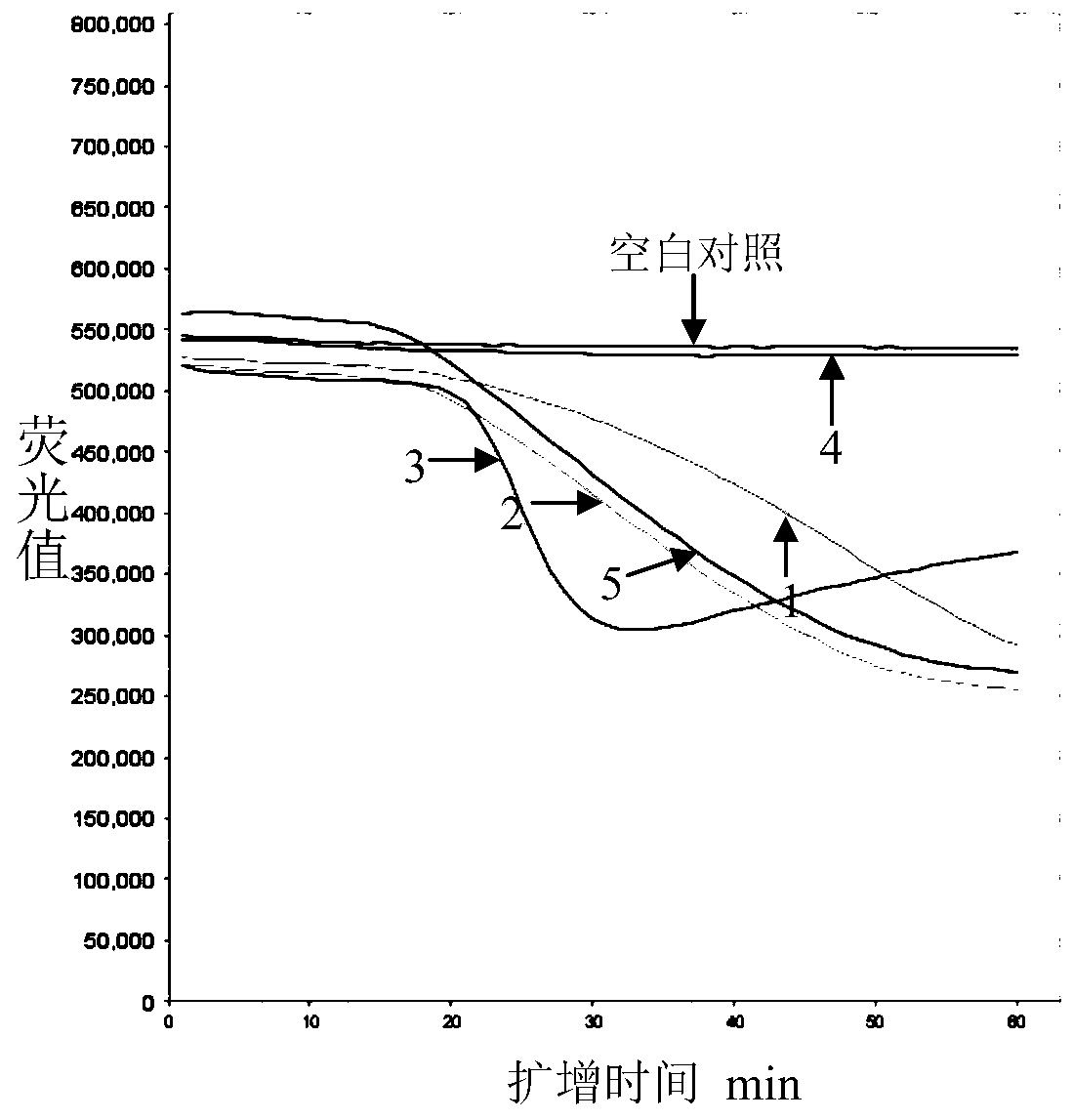
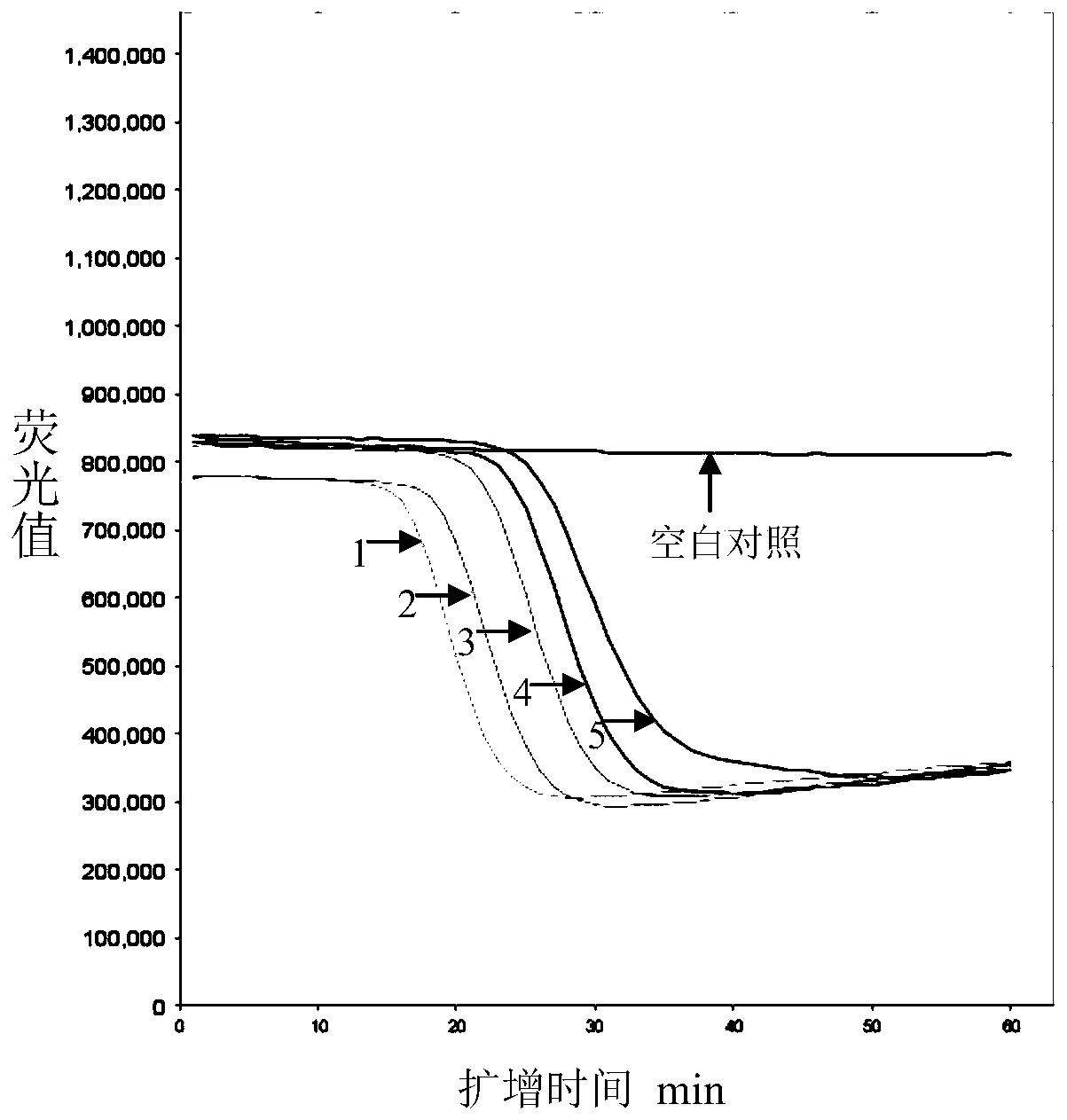
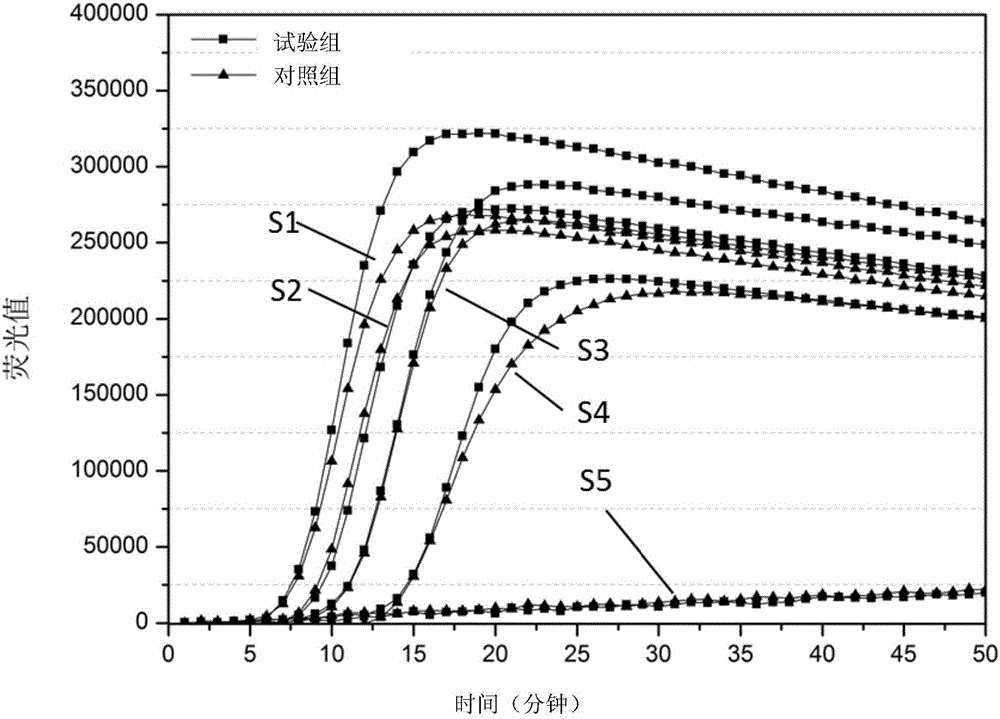
The invention relates to a method and kit for detecting a wild strain of a classical fever virus. The method for detecting the wild strain of the classical swine fever virus includes the following steps that (1), a detection sample is obtained, specifically, wild strain RNA of the classical swine fever virus is extracted, and the RNA concentration is diluted to 10 copies / [mu]L after extraction; (2), a reaction system is prepared, specifically, 7[mu]L of a constant temperature amplification buffer solution, 1[mu]L of a constant temperature amplification enzyme solution and 2[mu]L of a RNA solution diluted in the step (1) are taken and mixed into 10 [mu]L of a reaction solution, and the reaction solution is evenly mixed through vortex vibration; (3), nucleic acid sequence based amplification(NASBA) and detection are carried out, specifically, the reaction solution is put into a real-time fluorescent PCR instrument, an NASBA primer group and a probe group are used, the temperature is setto be 41 DEG C, constant temperature amplification reaction is carried out for 1h, and meanwhile, real-time fluorescent scanning is completed; and (4) a result is judged, specifically, when a typicallying-S type amplification curve is detected, the detecting result is positive.
Owner:BEIJING UNIV OF AGRI +1
Method for amplifying nucleic acid sequence
The present invention provides a convenient and effective method for amplifying a nucleic acid sequence characterized by effecting a DNA synthesis reaction in the presence of chimeric oligonucleotide primers; a method for supplying a large amount of DNA amplification fragments; an effective method for amplifying a nucleic acid sequence by combining the above method with another nucleic acid sequence amplification method; a method for detecting a nucleic acid sequence for detecting or quantitating a microorganism such as a virus, a bacterium, a fungus or a yeast; and a method for detecting a DNA amplification fragment obtained by the above method in situ.
Owner:TAKARA HOLDINGS
Nucleic acid amplification method
InactiveCN103667252AAvoid efficiencyAvoid featuresMicrobiological testing/measurementDNA preparationA-DNASingle strand dna
The invention belongs to the technical field of biology, and discloses a nucleic acid amplification method and a kit thereof. The method is characterized in that restriction enzyme is additionally utilized for amplifying double-chain DNA (Desoxvribose Nucleic Acid) and / or single-chain DNA, and other reagents are not added, so that the influence of the substances on the follow-up TMA (Thyroid Microsomal Antibody) or NASBA (Nucleic Acid Sequence Based Amplification) reaction is avoided, and as a result, the problems of reduction of amplification efficiency and increase of nonspecific products caused by additionally added restrictive oligonucleotides can be avoided. The method and the kit thereof are applied to DNA of any type, virus DNA to be detected in a sample, and genome DNA, and can be used for multiple detection of a DNA mixed sample as well as multiple detection of DNA and RNA (Ribose Nucleic Acid) mixed samples. The method can be widely used in match with other relevant technologies in the fields of molecular diagnosis, scientific research, epidemic prevention detection, medicolegal expertise and other fields.
Owner:SNOVA BIOTECH
Kit capable of detecting natural water sample of prorocentrum donghaiense in high throughput manner
The invention discloses a kit capable of detecting a natural water sample of prorocentrum donghaiense in a high-throughput manner. The kit comprises 12 constituents, including a natural sample nucleic acid crude-extract reagent, 5*NASBA (Nucleic Acid Sequence Based Amplification) Buffer, 5*Primer mix, 4*Enzyme mix, an RNA (Ribonucleic Acid) denaturing agent and a hybridization solution. The detection principle of the kit is as follows: rRNA (Ribosomal Robonucleic Acid) of an algae cell is taken as a target sequence which is prepared by using an isothermal nucleic acid amplification technology, and a plurality of detection samples are detected at the same time by virtue of the reverse dot blot of RNA. According to the kit, a detection process is in need of simple equipment only, the detection is rapid, a detection result can be judged directly through naked eyes, and the kit has high sensitivity, can be used for site detection of a daily water environment and has a certain practical value for early warning of the red tide of the prorocentrum donghaiense.
Owner:HARBIN INST OF TECH AT WEIHAI
Detection method and kit for transgenic soybean A2704-12 transformation event
InactiveCN102634595AReduce testing costsReduce mutual contaminationMicrobiological testing/measurementPolymerase LNucleic acid sequence based amplification
Owner:海康生物科技(北京)有限公司 +1
Nucleic acid encapsulating reagent and application thereof
ActiveCN106754900AGood embedding effectImprove adhesionBioreactor/fermenter combinationsBiological substance pretreatmentsZika virusDisease
The invention discloses a nucleic acid encapsulating reagent and an application thereof. The nucleic acid encapsulating reagent provided by the invention consists of 3-10 parts by mass of agarose, 1-5 parts by mass of saponin and 2-6 parts by mass of hydroxy propyl cellulose. The invention also discloses a target material detection device, wherein the target material detection device is obtained by embedding primers and a probe for detecting a target material in a reaction device by virtue of the nucleic acid encapsulating reagent. The invention also discloses a Zika virus detection device, wherein the Zika virus detection device is obtained by embedding a primer ZIKV-F as shown in a sequence 1, a primer ZIKV-R as shown in a sequence 2 and a probe ZIKV-P as shown in a sequence 3 in the reaction device by virtue of the nucleic acid encapsulating reagent prescribed in any one of previous claims. The invention has an important application value for detecting the target material with the application of a nucleic acid sequence-based amplification technology. The invention has great application and popularization value for detection of Zika viruses and for prevention and control of related diseases.
Owner:CAPITALBIO CORP
Method and kit for detecting food-borne pathogenic bacteria based on nasba test strips
ActiveCN103146835BImprove featuresHigh sensitivityMicrobiological testing/measurementMicroorganism based processesMicrobiologyNucleic acid sequence based amplification
The invention discloses a method and a kit for detecting pathogenic bacterium of a food source by a test strip based on NASBA (Nucleic Acid Sequence Based Amplification) and belongs to the technical field of biological detection. The method comprises the following steps of: (1) extracting RNA of pathogenic bacterium of the food source; (2) designing an amplification primer; (3) performing NASBA reaction; (4) designing a catching probe; (5) preparing a nano-gold probe; (6) preparing a collaurum nucleic acid strip; and (7) detecting the samples. The kit using the method to detect Listeria monocytogenes comprises primers (as shown in SEQ ID NO. 1 and 2), enzyme, buffer solution, RNaseH, RNA enzyme inhibitor, dNTPs, NTPs, probes (as shown in SEQ ID NO. 4, 5 and 6) and collaurum nucleic acid strip, etc. By combining NASBA with detection of the test strip, the method disclosed by the invention can realize qualitative or quantitative detection, and has low cost, high detection speed, good specificity, high sensitivity and safety in use.
Owner:SOUTH CHINA NORMAL UNIVERSITY
Primers, probes and kits for nasba-elisa detection of type Ⅱ grass carp reovirus
InactiveCN104388588BShort detection time periodImprove detection efficiencyMicrobiological testing/measurementMicroorganism based processesPositive controlSorbent
The invention discloses an NASBA-ELISA (nucleic acid sequence-based amplification-enzyme-linked immuno sorbent assay) detection primer, a probe and a kit for type II grass carp reovirus. Nucleotide sequences of the primer and the probe are as shown in SEQ ID NO.1-4. The kit contains the primer, the probe, an NASBA amplification buffer solution, an enzyme mixed liquor, an amplification product detection reagent, a negative control and a positive control. The kit for the type II grass carp reovirus has the advantages that a detection time period is short, and detection efficiency is high; virus detection specificity is high, and accuracy is high; virus quantitative analysis can be realized while virus qualitative analysis is realized; detection sensitivity is higher than that of common PCR (polymerase chain reaction); operation is easy, and popularization is easy; and repeatability of experimental results is good.
Owner:PEARL RIVER FISHERY RES INST CHINESE ACAD OF FISHERY SCI
Method for amplifying nucleic acid sequence
The present invention provides a convenient and effective method for amplifying a nucleic acid sequence characterized by effecting a DNA synthesis reaction in the presence of chimeric oligonucleotide primers; a method for supplying a large amount of DNA amplification fragments; an effective method for amplifying a nucleic acid sequence by combining the above method with another nucleic acid sequence amplification method; a method for detecting a nucleic acid sequence for detecting or quantitating a microorganism such as a virus, a bacterium, a fungus or a yeast; and a method for detecting a DNA amplification fragment obtained by the above method in situ.
Owner:TAKARA HOLDINGS
NASBA-ELISA (Nucleic Acid Sequence Based Amplification-Enzyme-Linked Immuno Sorbent Assay) detection primer and probe for detecting porcine epidemic diarrhea
InactiveCN112011536ARapid diagnosisStrong specificityMicrobiological testing/measurementMaterial analysisDiseaseNucleotide
The invention relates to an NASBA-ELISA (Nucleic Acid Sequence Based Amplification-Enzyme-Linked Immuno Sorbent Assay) detection primer and probe for detecting porcine epidemic diarrhea. According toan N gene segment of a variant porcine epidemic diarrhea virus, a synthetic primer and a biotin probe are designed, DIG-11-UTP nucleotide is adopted to replace a digoxin detection probe, and an NASBA-ELISA detection method is established. The NASBA-ELISA detection method established by adopting the primer and the probe has no cross reaction with porcine reproductive and respiratory syndrome virus,swine fever virus, transmissible gastroenteritis virus, porcine circovirus type 2 and porcine pseudorabies virus, 4.3 pg of virus nucleic acid can be detected, and good specificity, sensitivity and repeatability are achieved. The method disclosed by the invention is large in one-time detection amount, does not need special expensive instruments and high-requirement technical content, is suitablefor rapid diagnosis of the porcine epidemic diarrhea disease by grassroots employees, and has important significance for effectively diagnosing, preventing and controlling the porcine epidemic diarrhea virus.
Owner:INST OF ANIMAL HUSBANDRY & VETERINARY FUJIAN ACADEMY OF AGRI SCI
Method for inspecting nucleic acid signal amplification of ligation nucleic acid intrusive reaction and cutting endonuclease reaction
InactiveCN101974638BLow costNo cross contaminationMicrobiological testing/measurementFluorescence/phosphorescenceFluorescenceNucleic acid sequencing
The invention discloses a method for detecting nucleic acid signal amplification of ligation nucleic acid intrusive reaction and cutting endonuclease reaction, which belongs to the field of molecular biology. The method comprises the following steps: 1) during nucleic acid intrusive signal amplification, accumulating flap fragments; 2) during flap fragment connection, connecting the flap fragments with another oligonucleotide on a molecular beacon to form a cutting endonuclease site; 3) during cutting endonuclease signal amplification, only cutting a molecular beacon chain so that the fluorescent group of the molecular beacon is separated from a quenching group, and a new integrated molecular beacon is hybridized with a flap fragment connected with the molecular beacon, thus amplifying a target nucleic acid signal; and 4) inspecting the target nucleic acid signals. The method is capable of inspecting nucleic acid target sequences and respectively inspecting nucleic acid sequences with single base difference near intrusive sites.
Owner:HUADONG RES INST FOR MEDICINE & BIOTECHNICS
Novel gene quantifying method
InactiveCN102517397ASimple calculationWide applicabilityMicrobiological testing/measurementNucleic acid sequence based amplificationBioinformatics
The invention provides a novel gene quantifying method, which comprises the following steps of: (1) verifying the sensitivity of a group of amplification reagents through an experiment to detect a lowest gene quantity; (2) diluting templates to be quantified by different times and performing gene amplification on the templates of different concentrations with the same group of amplification reagents; and (3) calculating the quantity of genes to be quantified through a maximum diluting time which can be used for realizing gene amplification. In the method, only original gene amplification equipment is required, so that equipment investment is simplified greatly, the cost is low, and the method is suitable for basic level and field use; only one multiplication calculation is applied, and the multiplication calculation is simple and easy to learn; and the method has wide suitability, and can be combined with the conventional gene amplification technologies such as PCR (Polymerase Chain Reaction), LAMP (Loop-Mediated Isotheral Amplification), NASBA (Nucleic Acid Sequence Based Amplification) and the like for completing a quantifying process.
Owner:李治国 +1
Detection of nucleic acid sequence modification
InactiveUS20100291695A1Precise temperature regulationLevel of detectionMicrobiological testing/measurementBiological testingTemperature controlNucleotide
The invention relates to a non-PCR based method for the detection of a nucleotide modification in a nucleic acid sample comprising contacting a solution comprising nucleic acid sample with a nucleic acid probe in a temperature-controlled and UV illuminated container and measuring the UV absorption of the nucleic acid sample / nucleic acid probe complex.
Owner:DELTADOT
A kind of highly sensitive respiratory virus nucleic acid nasba amplification primer and detection method
ActiveCN105256068BHigh detection sensitivityEliminate hazardsMicrobiological testing/measurementDNA/RNA fragmentationThroat swab sampleT7 RNA polymerase
The invention provides novel high-sensitivity respiratory virus nucleic acid NASBA (nucleic acid sequence based amplification) primers. A respiratory virus conservative area is selected, and a promoter sequence capable of being recognized by T7 RNA (ribose nucleic acid) polymerase is formed at the 5' terminal of one of the synthesized primers. The invention further provides a novel high-sensitivity respiratory virus nucleic acid NASBA detection method. T7 RNA polymerase promoter sequences are added to the 5' terminals of the two primers complementary to two ends of a respiratory virus RNA sequence, so that when the primers enter a circulation phase, RNA sequences [RNA(+)] consistent with a template sequence can be synthesized, RNA sequences [RNA(-)] complementary to a template RNA sequence can be synthesized, and the detection sensitivity of viral nucleic acid is greatly improved; a throat swab sample of a patient can be directly used for amplification, a reverse transcription process is not required, and the operation time is saved.
Owner:HANGZHOU JOINSTAR BIOTECH
Nasba amplification primers, kits and detection methods for detecting hop dwarf viroid
InactiveCN104450973BGuaranteed reliabilityReduce the number of cyclesMicrobiological testing/measurementDNA/RNA fragmentationElectrophoresesDisease surveillance
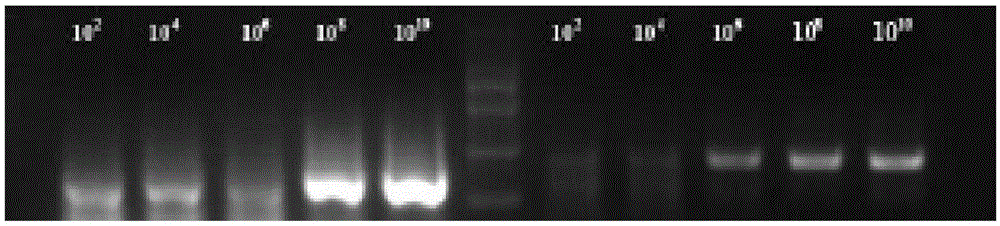
The invention discloses an NASBA (nucleic acid sequence based amplification) primer, a kit and a detection method for detecting hop stunt viroids. The sequences of the upstream primer and the downstream primer of the NASBA primer are SEQ ID NO: 1-2 respectively, and the kit comprises NASBA reaction liquid A and NASBA reaction liquid B. The detection method includes the steps: 1) extracting RNA (ribonucleic acid) of samples; 2) performing NASBA for the hop stunt viroids; 3) detecting electrophoreses. The NASBA primer is applicable to rapidly detecting and confirming the hop stunt viroids, can be widely applied to disease surveillance and control in agricultural production and environments and monitoring and detecting of the viroids in import and export trade and is simple and convenient to operate, the number of the needed samples is small, and the quality requirement for template RNA is low.
Owner:INSPECTION & QUARANTINE TECH CENT SHANDONG ENTRY EXIT INSPECTION & QUARANTINE BUREAU
A kind of nucleic acid encapsulation reagent and its application
ActiveCN106754900BEasy to operateSolve pollutionBioreactor/fermenter combinationsBiological substance pretreatmentsZika virusNucleic acid sequence based amplification
The invention discloses a nucleic acid encapsulating reagent and an application thereof. The nucleic acid encapsulating reagent provided by the invention consists of 3-10 parts by mass of agarose, 1-5 parts by mass of saponin and 2-6 parts by mass of hydroxy propyl cellulose. The invention also discloses a target material detection device, wherein the target material detection device is obtained by embedding primers and a probe for detecting a target material in a reaction device by virtue of the nucleic acid encapsulating reagent. The invention also discloses a Zika virus detection device, wherein the Zika virus detection device is obtained by embedding a primer ZIKV-F as shown in a sequence 1, a primer ZIKV-R as shown in a sequence 2 and a probe ZIKV-P as shown in a sequence 3 in the reaction device by virtue of the nucleic acid encapsulating reagent prescribed in any one of previous claims. The invention has an important application value for detecting the target material with the application of a nucleic acid sequence-based amplification technology. The invention has great application and popularization value for detection of Zika viruses and for prevention and control of related diseases.
Owner:CAPITALBIO CORP
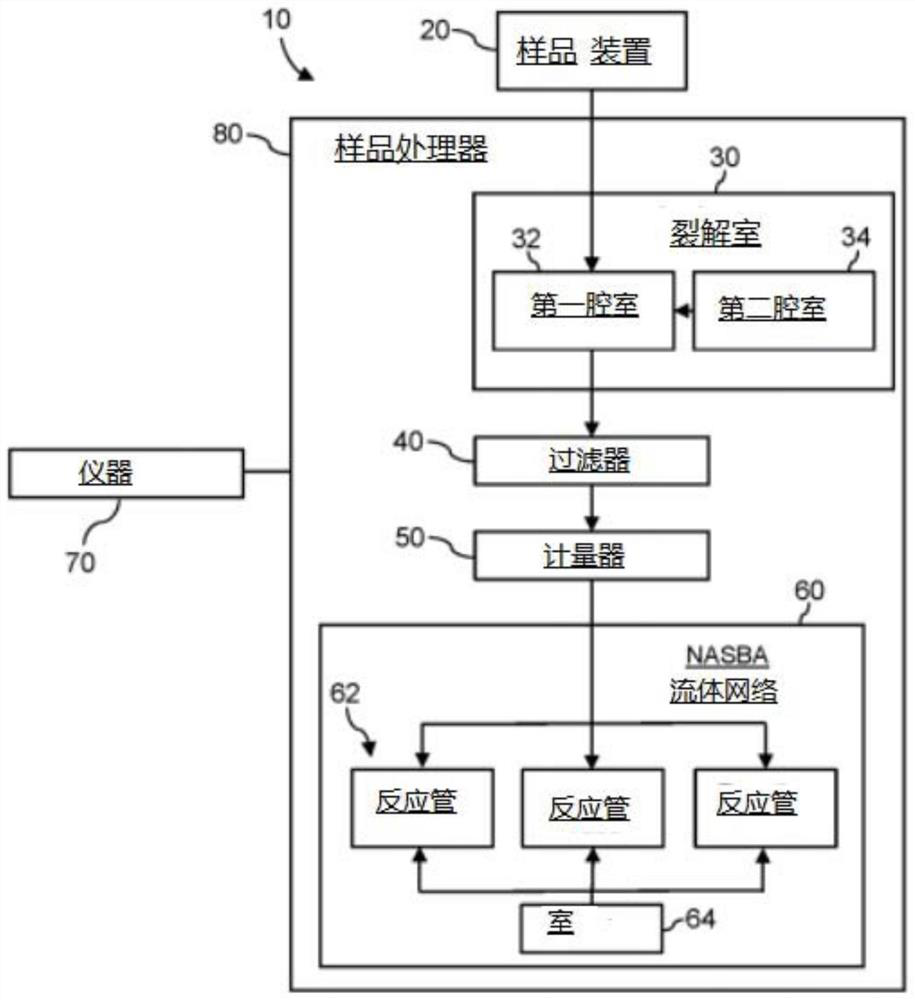
Method for detecting presence of severe acute respiratory syndrome coronavirus 2 in biological sample
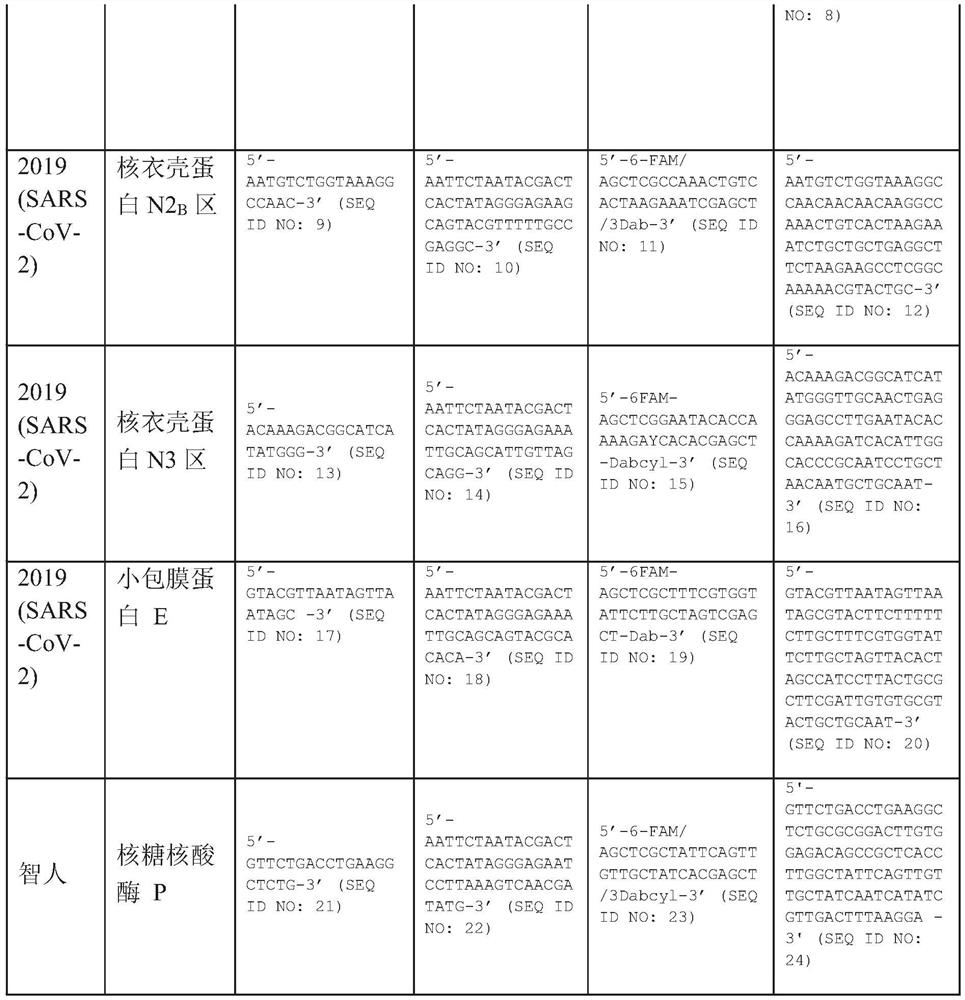
The present invention relates to a method for detecting the presence of SARS-CoV-2 in a biological sample, comprising: subjecting the biological sample to cleavage to form a lysate; generating an amplified lysate by nucleic acid sequence (NASBA)-based amplification of a target nucleic acid sequence in the lysate in the presence of a forward primer having an oligonucleotide sequence selected from the group consisting of SEQ ID NO: 1, SEQ ID NO: 5, SEQ ID NO: 9, SEQ ID NO: 13 and SEQ ID NO: 17, a reverse primer, and a molecular beacon; the reverse primer has an oligonucleotide sequence selected from the group consisting of SEQ ID NO: 2, SEQ ID NO: 6, SEQ ID NO: 10, SEQ ID NO: 14 and SEQ ID NO: 18; the molecular beacon has an oligonucleotide sequence selected from the group consisting of SEQ ID NO: 3, SEQ ID NO: 7, SEQ ID NO: 11, SEQ ID NO: 15, and SEQ ID NO: 19, and a fluorophore; exposing the amplified lysate to an excitation source; detecting fluorescence of a fluorophore in the amplified lysate exposed to the excitation source; in response to detecting the fluorescence of the fluorophore, it is determined whether SARS-CoV-2 is present in the biological sample.
Owner:TELEFLEX MEDICAL INC
Detection method and kit for transgenic soybean A2704-12 transformation event
InactiveCN102634595BReduce testing costsReduce mutual contaminationMicrobiological testing/measurementPolymerase LNucleic acid sequence based amplification
The invention discloses a method and a kit for rapidly detecting a transgenic soybean A2704-12 transformation event. The method comprises the following steps of: performing high-temperature denaturation on a sample DNA (deoxyribonucleic acid), and performing transcription by using a primer for identifying an A2704-12 specificity region under the action of RNA (ribonucleic acid) polymerase to produce RNA; taking the obtained RNA as a template, and performing NASBA (nucleic acid sequence based amplification) by using the primer; and finally detecting the amplified product. The method does not need to purchase a large instrument and thus reduces the detection cost; and the detected product is easy to degrade, thus reducing the problem of cross contamination among samples.
Owner:海康生物科技(北京)有限公司 +1
A highly specific probe for real-time detection of nasba products
ActiveCN106048014BHigh sensitivityStrong specificityMicrobiological testing/measurementDNA/RNA fragmentationNucleic acid amplification techniqueTrue positive rate
The invention belongs to the technical field of nucleic acid amplification, and relates to a highly specific probe for real-time detection of NASBA (Nucleic Acid Sequence Based Amplification) products and a method for detecting the NASBA products. The highly specific probe comprises two adjacent nucleotide sequences, bases from the first position to the fourth position at the 3' end of each of the two adjacent nucleotide sequences are ribonucleotide bases, and the remaining bases are all deoxyribonucleotide bases. The highly specific probe is applied to detection of the NASBA products, has high sensitivity and specificity, and reduces detection time, thereby enabling the NASBA detection technology to be better applied to detection of nucleic acid of pathogenic microorganisms.
Owner:CAPITALBIO CORP +1
A detection method and kit for swine fever virus based on the fluorescence characteristics of g-quadruplex
InactiveCN104611462BHigh amplification efficiencyHigh detection sensitivityMicrobiological testing/measurementMicroorganism based processesSwine Fever VirusVirus detection
The invention discloses a swine fever virus detection method based on G-quadruplex fluorescence characteristic and a kit, and belongs to the technical field of gene engineering. The method comprises: utilizing a nucleic acid sequence-based amplification (NASBA) technology to amplify target RNA to obtain a single-chain RNA product; adding an upstream probe, a downstream probe and the RNA amplification product into a G- quadruplex fluorescence detection buffer, performing denaturation and annealing, then adding protoporphyrin, and utilizing a microplate reader or fluorophotometer to detect the fluorescence intensity in the reaction system, so as to determine whether the sample possesses a swine fever virus. The method is fast, precise and stable in swine fever virus detection speed, good in reappearance, simple in detection steps and low in cost.
Owner:GUIZHOU DAXING AGRI SCI & TECH DEV CO LTD +1
Molecular detection and quantification of Enterococci
InactiveUS7947441B2Improve abilitiesEnsure resource securitySugar derivativesMicrobiological testing/measurementNucleic acid sequence based amplificationBioinformatics
A primer pair and probe for the large subunit ribosomal RNA gene of enterococci for use in a real-time nucleic acid sequence based amplification (NASBA) assay.
Owner:UNIV OF SOUTH FLORIDA
A kit for high-throughput detection of natural water samples of Prorocentrum donghaiense
InactiveCN103091311BEasy to detectMaterial analysis by observing effect on chemical indicatorRed tideBiology
The invention discloses a kit capable of detecting a natural water sample of prorocentrum donghaiense in a high-throughput manner. The kit comprises 12 constituents, including a natural sample nucleic acid crude-extract reagent, 5*NASBA (Nucleic Acid Sequence Based Amplification) Buffer, 5*Primer mix, 4*Enzyme mix, an RNA (Ribonucleic Acid) denaturing agent and a hybridization solution. The detection principle of the kit is as follows: rRNA (Ribosomal Robonucleic Acid) of an algae cell is taken as a target sequence which is prepared by using an isothermal nucleic acid amplification technology, and a plurality of detection samples are detected at the same time by virtue of the reverse dot blot of RNA. According to the kit, a detection process is in need of simple equipment only, the detection is rapid, a detection result can be judged directly through naked eyes, and the kit has high sensitivity, can be used for site detection of a daily water environment and has a certain practical value for early warning of the red tide of the prorocentrum donghaiense.
Owner:HARBIN INST OF TECH AT WEIHAI
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com