A highly specific probe for real-time detection of nasba products
A probe and specific technology, applied in the field of high-specific probes for real-time detection of NASBA products, can solve the problems of difficult diagnosis, non-specific reaction, high concentration, etc., and achieve the effect of shortening the detection time, strong specificity and high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
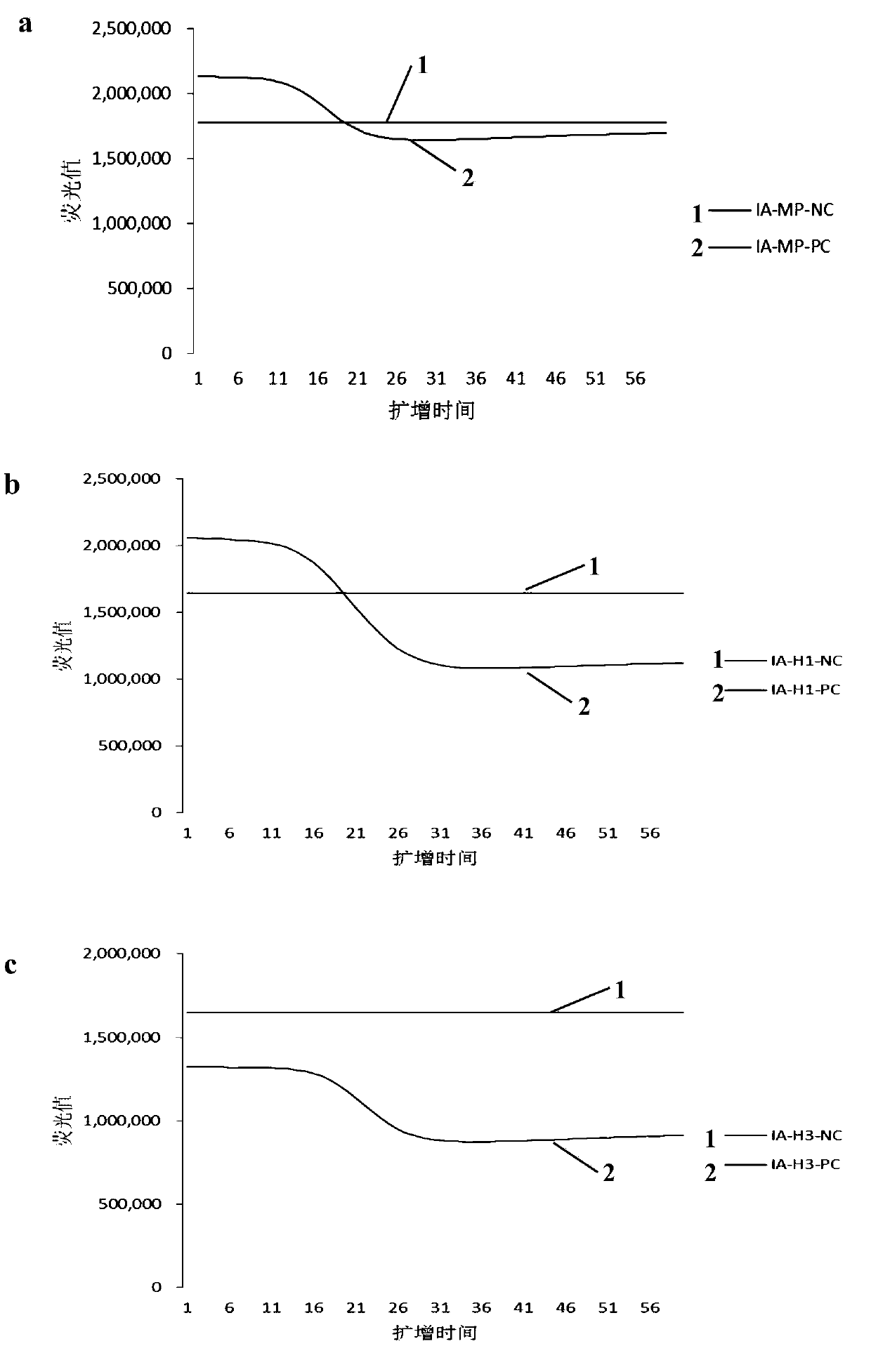
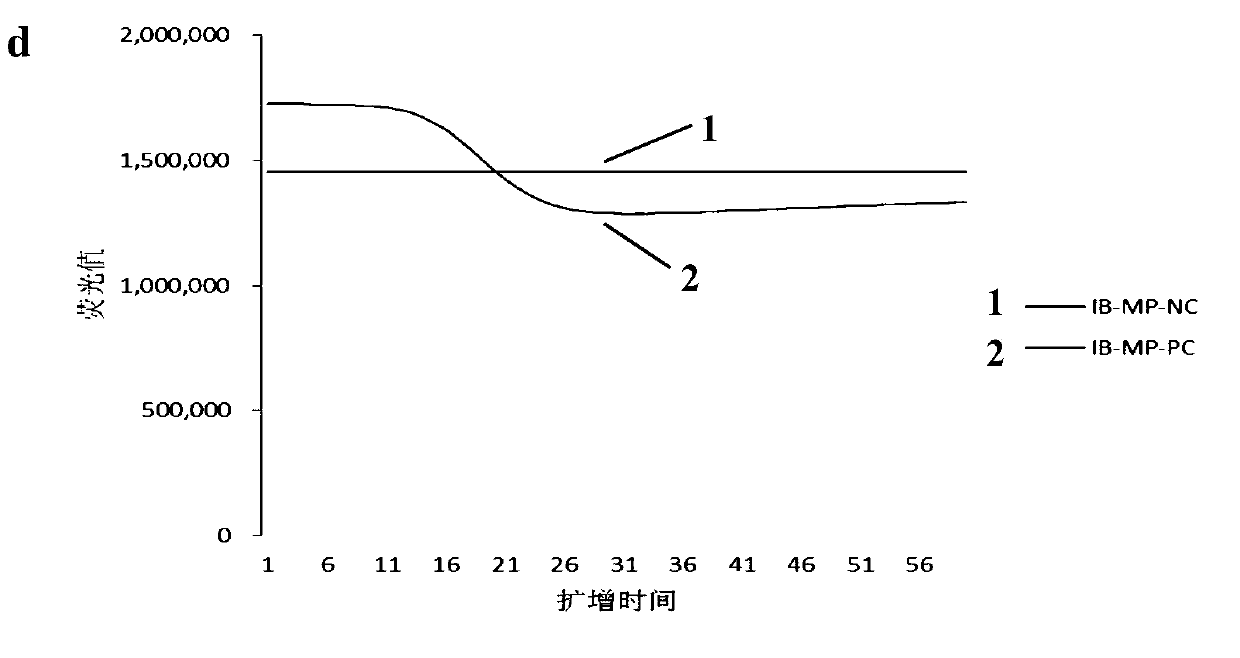
[0028]Embodiment 1, the adjacent probe that is used for the detection of influenza virus NASBA amplification product
[0029] The present invention designs four sets of primers and adjacent probes for the detection of influenza virus NASBA amplification products, the primers used for influenza A virus gene detection are InfA-TY-PrimerF, InfA-TY-PrimerR, and the corresponding adjacent probes are InfA -TY-ProbeF, InfA-TY-ProbeR, the primers used for the detection of influenza A virus H1 subtype gene are InfA-H1-PrimerF, InfA-H1-PrimerR, and the corresponding adjacent probes are InfA-H1-ProbeF, InfA- H1-ProbeR, the primers used for the detection of influenza A virus H3 subtype genes are InfA-H3-PrimerF, InfA-H3-PrimerR, and the corresponding adjacent probes are InfA-H3-ProbeF, InfA-H3-ProbeR, which are used for B The primers for influenza virus gene detection are InfB-TY-PrimerF, InfB-TY-PrimerR, and the corresponding adjacent probes are InfB-TY-ProbeF, InfB-TY-ProbeR. The NASBA...
Embodiment 2
[0035] Embodiment 2, adjacent probe is used for NASBA product detection
[0036] 1. Preparation of RNA template
[0037] The plasmids used to prepare RNA templates were provided by Boao Bio Group Co., Ltd., including recombinant plasmid pUC-InfA-TY containing influenza A virus gene, recombinant plasmid pUC-InfA-H1 containing influenza A virus H1 subtype HA gene, containing The recombinant plasmid pUC-InfA-H3 containing the HA gene of influenza A virus H3 subtype, and the recombinant plasmid pUC-InfB-TY containing the gene of influenza B virus. The RNA template preparation process is as follows.
[0038] 1. Enzyme digestion: first digest each recombinant plasmid to be tested with EcoRI endonuclease at 37°C for 2 hours.
[0039] 2. Transcription: Prepare a reaction with a total volume of 50 μL by 5 μL 5×Transcription Optimized Buffer (Promega), 2U T7 RNA polymerase, 10 mM DTT (Promega), 10 U recombinant RNase inhibitor (Promega), 2 mM rNTP, and 5 μL of digested product system...
Embodiment 3
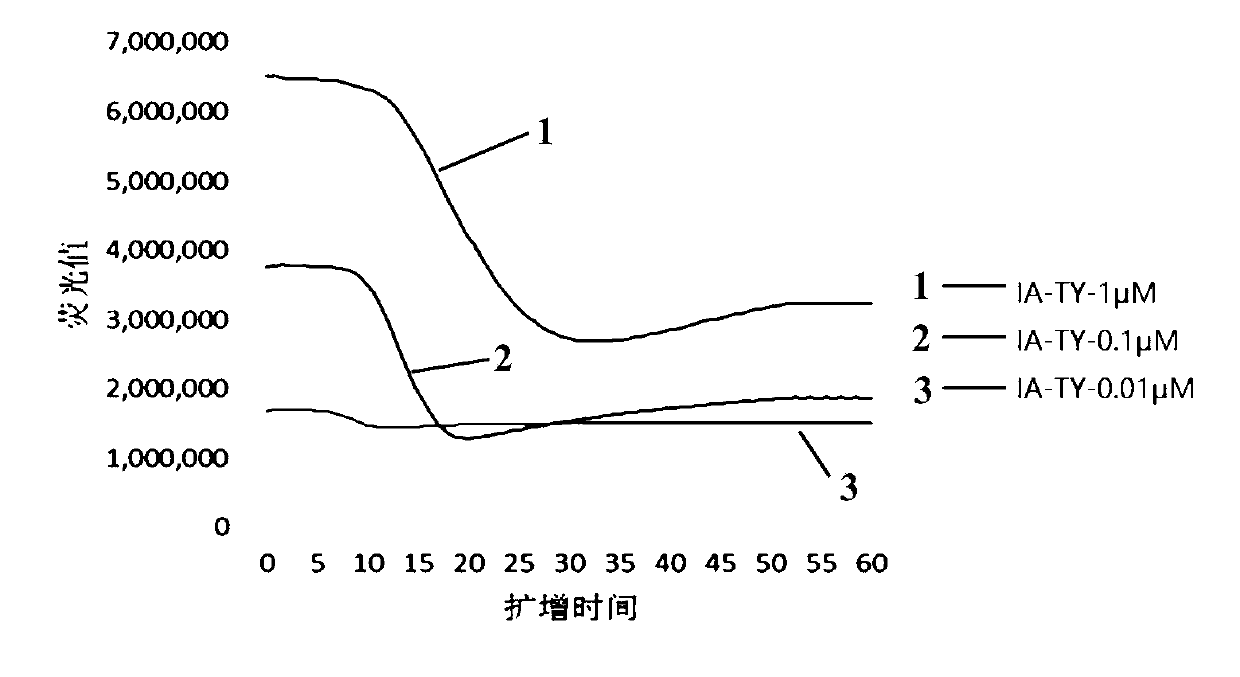
[0060] Embodiment 3, adjacent probe characteristic analysis
[0061] 1. Concentration of adjacent probes
[0062] 1. Preparation of constant temperature amplification system with different adjacent probe concentrations
[0063] Isothermal amplification buffer B1: 200mM Tris-HCL (pH 8.0), 0.5μM InfA-TY-PrimerF, 0.5μM InfA-TY-PrimerR, 0.01μM InfA-TY-ProbeF, 0.01μM InfA-TY-ProbeR, 50mM DTT, 10mM dNTP, 10mM rNTP, 80mM MgCl 2 , 450mM KCl, 15% by volume DMSO, 1M sorbitol, 20mM tetramethylammonium chloride.
[0064] Isothermal amplification buffer B2: 200mM Tris-HCL (pH 8.0), 0.5μM InfA-TY-PrimerF, 0.5μM InfA-TY-PrimerR, 0.1μM InfA-TY-ProbeF, 0.1μM InfA-TY-ProbeR, 50mM DTT, 10mM dNTPs, 10mM rNTPs, 80mM MgCl 2 , 450mM KCl, 15% by volume DMSO, 1M sorbitol, 20mM tetramethylammonium chloride.
[0065] Isothermal amplification buffer B3: 200mM Tris-HCL (pH 8.0), 0.5μM InfA-TY-PrimerF, 0.5μM InfA-TY-PrimerR, 1μM InfA-TY-ProbeF, 1μM InfA-TY-ProbeR, 50mM DTT, 10mM dNTP , 10mM rNTP, 80m...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com