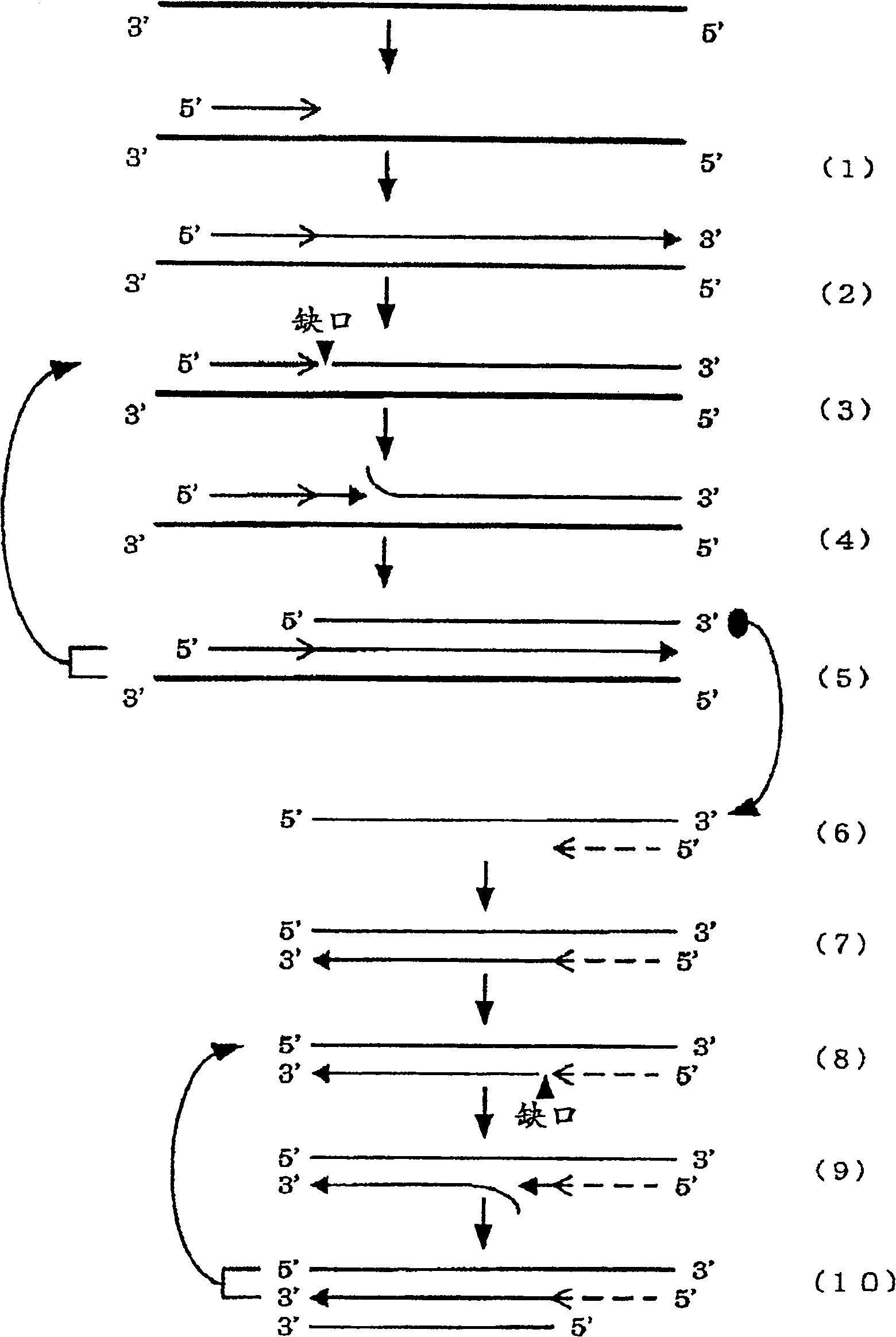
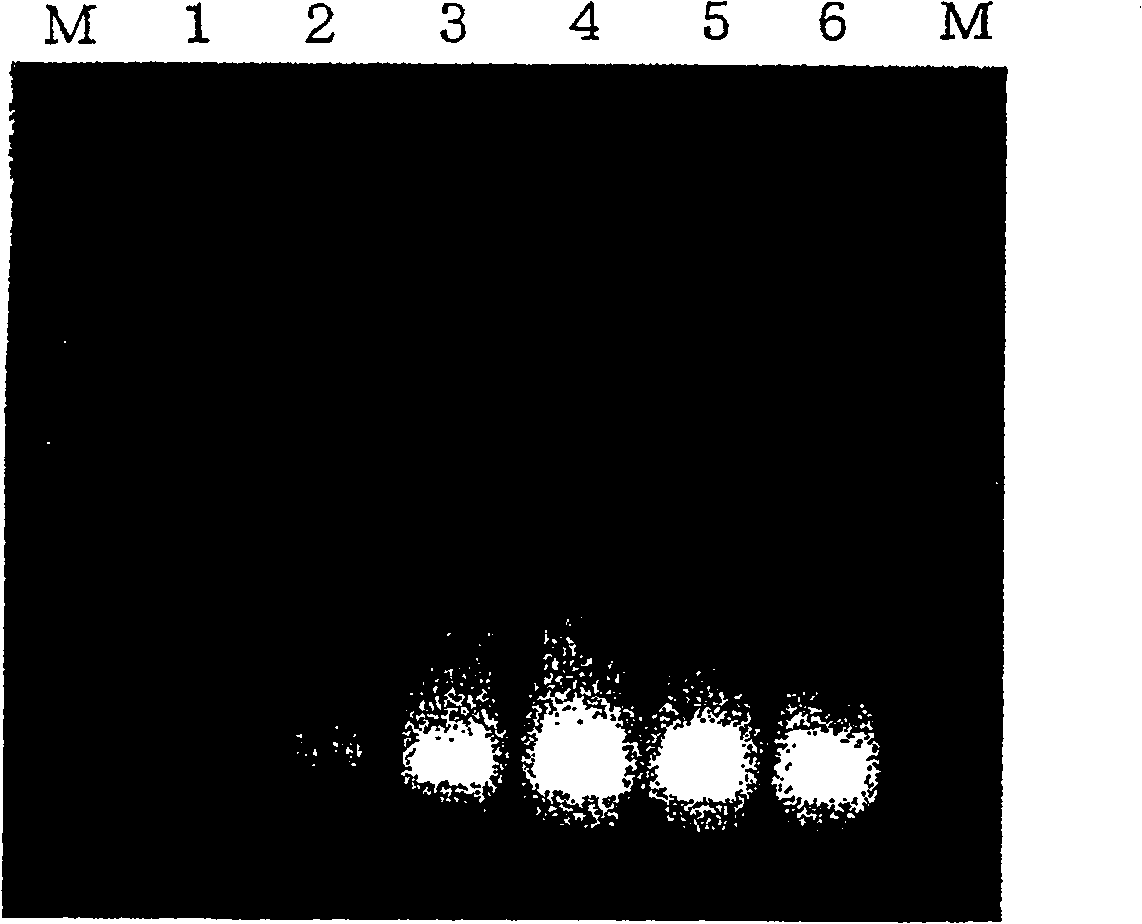
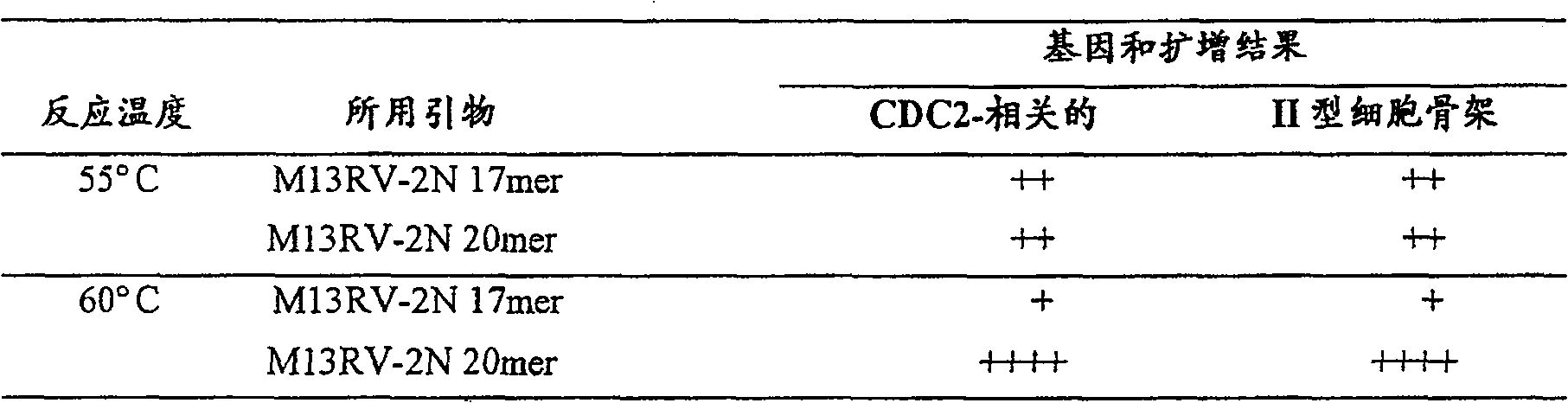
Method for amplifying nucleic acid sequence
A technology of nucleotide sequence and nucleic acid, applied in the field of synthetic DNA
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0259] (1) Synthetic template DNA and primers
[0260] A single-stranded DNA of 99 bases used as a template and primers used in this example were synthesized using a DNA synthesizer (Applied Biosystems). The nucleotide sequence of the 99-base single-stranded DNA is shown in SEQ ID NO: 1 in the Sequence Listing. The basic nucleotide sequences of the upstream primer and the downstream primer are respectively shown in SEQ ID NO: 2 and 3 of the sequence listing. The structure of the primers used in this embodiment is described in detail below:
[0261] Primer pair 1: a combination of primers having the nucleotide sequence shown in SEQ ID NO: 2 or 3 of the sequence table and all consisting of deoxyribonucleotides;
[0262] Primer pair 2: wherein in each primer of primer pair 1, the first and second deoxyribonucleotides at the 3'-terminus are replaced with ribonucleotides and the second ribonucleoside at the 3'-terminus A combination of primers in which the phosphate bond on the ...
Embodiment 2
[0280] (1) Preparation of RNA
[0281] RNA used as a template in this example was prepared from cultured human cells HT29 (ATCC HTB-38) (Dainippon Pharmaceutical) using TRIzol reagent (Life Technologies). The concentration of the obtained total RNA was adjusted to 1 µg / µl. The OD260 / OD280 value is 1.8, which indicates the spectrophotometric purity of the RNA.
[0282] (2) Amplification reaction
[0283] Bca BEST DNA polymerase with reverse transcription activity and DNA polymerase activity and RNase H endonuclease were used to determine whether cDNA was amplified from RNA.
[0284] A reaction mixture having the composition described in Example 2 was prepared by adding 1 μg of the above total RNA. The target region encoding human transferrin receptor (gene accession number X01060) was amplified using primer pair 2 of Example 1 as primers.
[0285] The reaction mixture was incubated at 55°C for 60 minutes, then heated at 90°C for 2 minutes to inactivate the enzyme. When 8 μ...
Embodiment 3
[0287] (1) Synthetic primers
[0288] The amplification method of the present invention was studied using double-stranded DNA as a template. The primers used were synthesized with a DNA synthesizer (Applied Biosystems). The basic nucleotide sequences of the primers are shown in SEQ ID NO: 5-13 of the Sequence Listing. The structures of the primers used in this example are described in detail below. pUC19 DNA (Takara Shuzo) was used as a template for primer pairs A-F. The nucleotide sequence of pUC19 can be obtained from a database (gene accession number L09137). The amplified double-stranded DNA fragment was used as a template for primer pair G. According to the attached standard method, primers and TaKaRa RNA PCR kit (AMV) Ver.2.1 (Takara Shuzo) with the sequence shown in SEQ ID NO: 14 or 15 of the sequence listing were used to prepare from the human total RNA obtained in Example 2 fragment.
[0289] Primer pair A (length of amplified fragment: about 450bp): having the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com