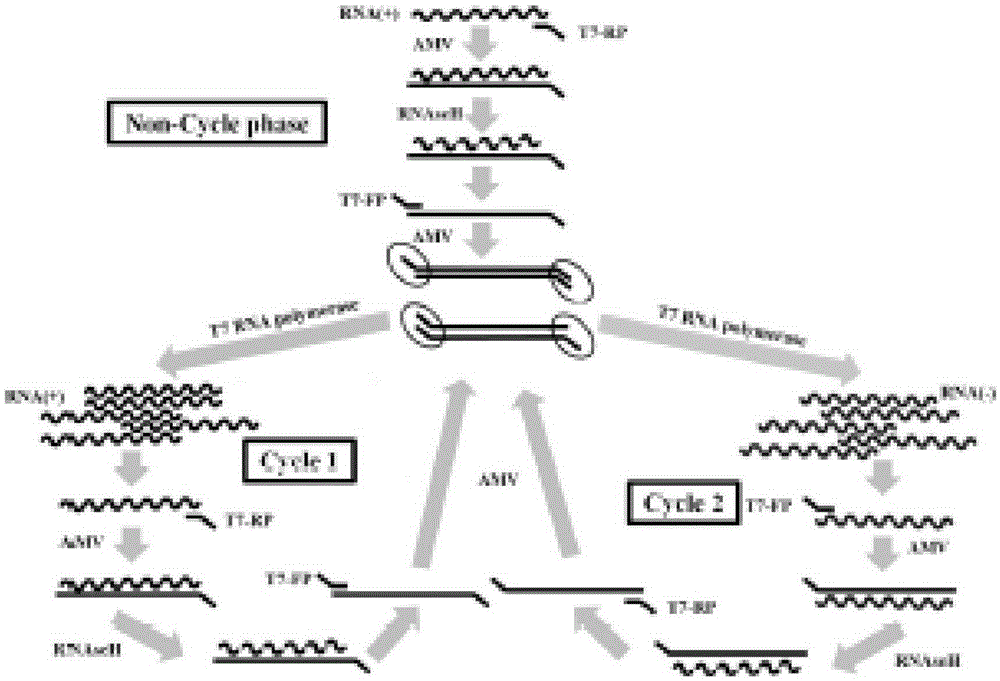
Novel high-sensitivity respiratory virus nucleic acid NASBA (nucleic acid sequence based amplification) primers and detection method
An amplification primer and high-sensitivity technology, applied in the field of biomedicine, can solve the problems of increased pollution opportunities, long time-consuming, multiple scientific research purposes, etc., and achieve the effects of improving RNA amplification efficiency, improving detection sensitivity, and saving operation time.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0081] Reagent composition (taking respiratory syncytial virus as an example)
[0082] (1) Primer
[0083] FP: AATTCTAATACGACTCACTATAGGGAGAAGGAAAGTCCTACAAAAAAATGC
[0084] RP: AATTCTAATACGACTCACTATAGGGAGAAGGATCTATCTCCTGCTGCTAAT 0.25ul each.
[0085] (2) NASBA Amplification Reagent
[0086] BA (buffer): 2ul
[0087] DA:dNTP0.25ul
[0088] NA: NTP0.5ul
[0089] DMSO: 0.25ul
[0090] EM (enzyme mix): AMV, RNAseH, T7 RNA polymerase, BSA5.5ul
[0091] (3) NASBA product detection reagent
[0092] 1% agarose gel
Embodiment 2
[0093] Embodiment 2: NASBA detection process
[0094] (1) Amplification system: Add 1ul sample RNA template, 2ulBA, 0.25ulNA, 0.5ulDA, 0.25ulDMSO, 0.25ulFP, 0.25ulRP into a 0.2ml PCR amplification tube.
[0095] (2) Amplification process: Place the PCR amplification tube on the amplification instrument, heat it at 65° for 5 minutes, then cool it at 42° for 5 minutes; then add 5.5ulEM, and react at 42° for 60 minutes.
[0096] (3) Amplification product detection: Take 2ul of sample buffer and add it to the amplification product to terminate the reaction.
[0097] The amplified product was added to 1% agarose gel, and electrophoresis standard was added to one of the sample wells. After electrophoresis at 100V for 10 minutes, the presence or absence of the target band was observed in the electrophoresis imager.
Embodiment 3
[0098] Embodiment 3: comparative experiment
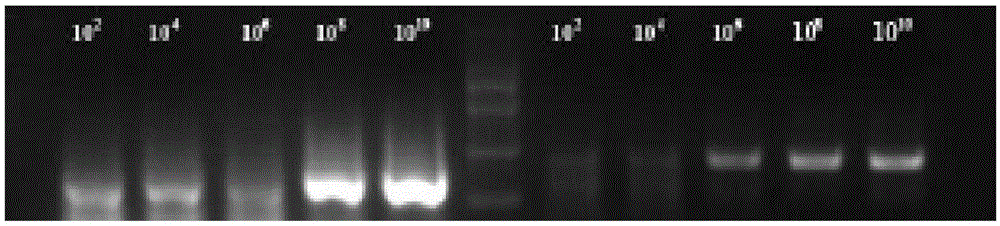
[0099] Adopt the contrast of respiratory syncytial virus NASBA kit of the present invention and one-step PCR method to dilute the secondary standard of respiratory syncytial virus into 10 2 、10 4 -10 10 copy / ul. Use the kit of the present invention and one-step PCR to detect simultaneously.
[0100] (1) NASBA kit sensitivity test
[0101] NASBA amplification and detection were performed according to Example 2.
[0102] From attached figure 2 It can be seen that the NASBA kit of the present invention can successfully detect 10 2 Copy the sample of / ul.
[0103] (2) One-step PCR sensitivity and rigidity detection
[0104] A. One-step PCR reaction system: 2ul sample RNA template, 12.5ul2*one-stepSYBRRT-PCRbuffer, 1ulprimeScript1stepEnzymeMix, 1ulFP, 1ulrp, 7.5ulRNaseFreeddh 2 0.
[0105] B. One-step PCR reaction process: put the above PCR tube at 42° for 5 minutes and then enter the cycle without going, 95° for 10 seconds,...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com