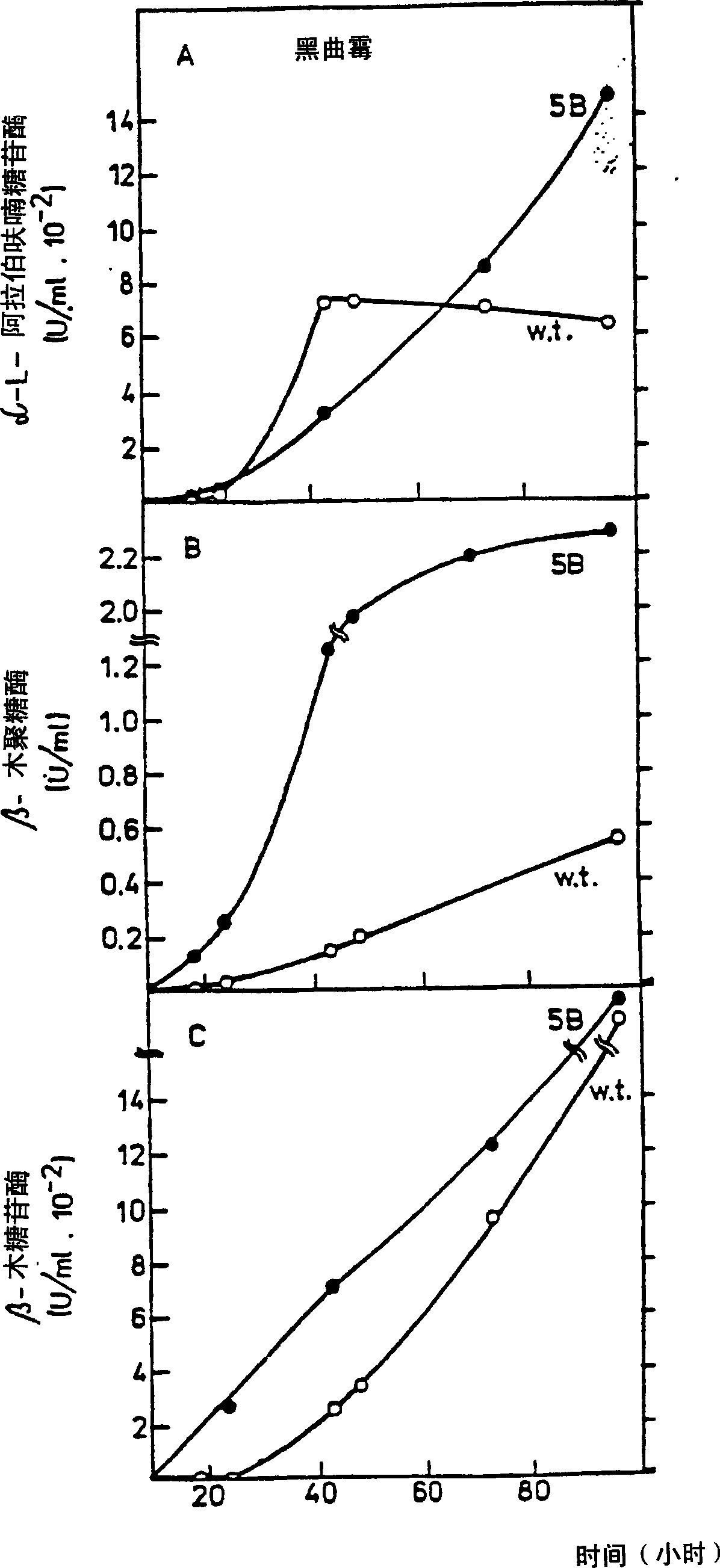
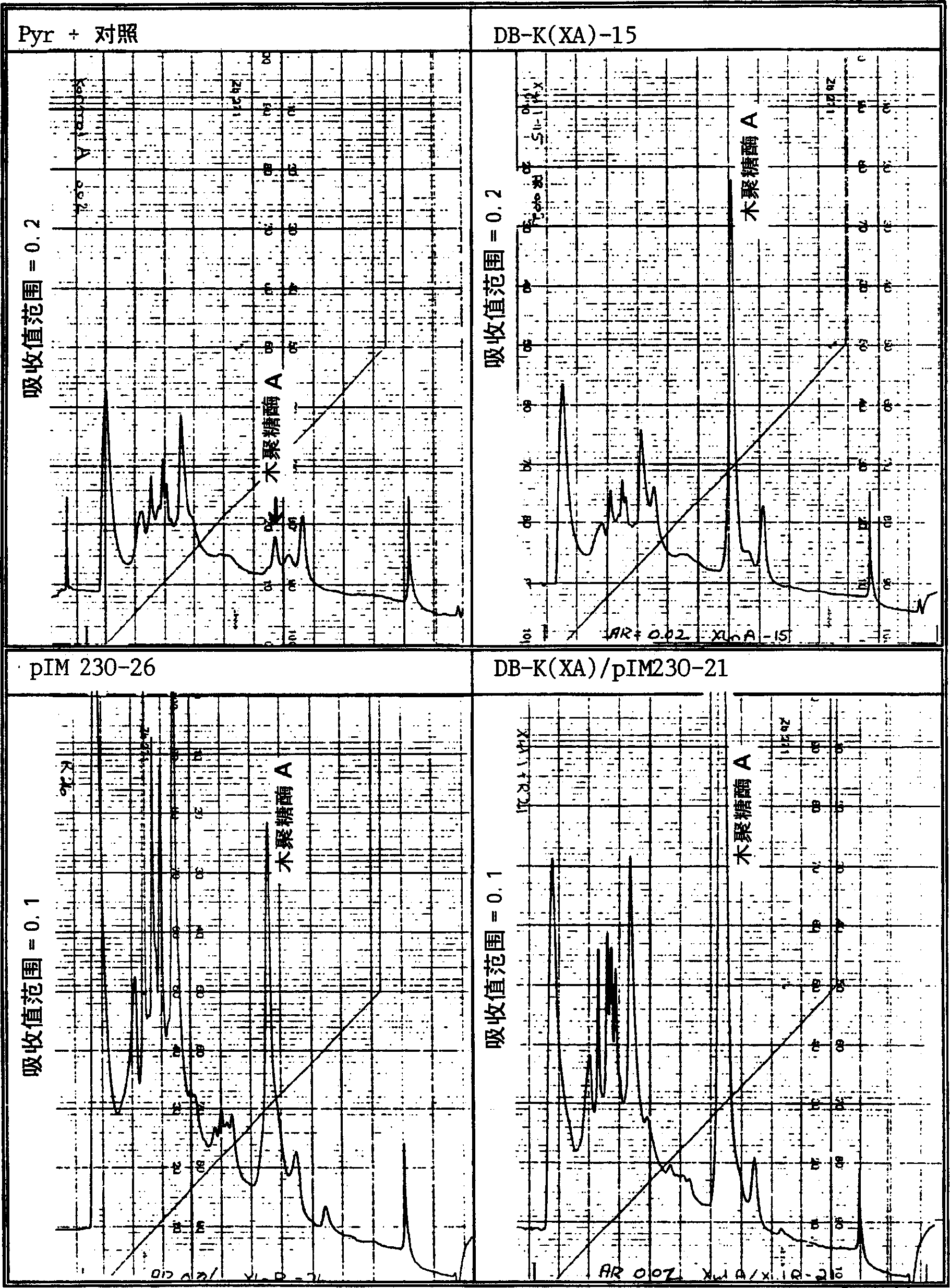
Novel method to isolate mutants and to clone the complementing gene
A target gene and amino acid technology, applied in the field of microbial mutation and screening of mutants, can solve complex time-consuming, non-specific binding and other problems
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0120] Example 1: Construction of plasmids
Embodiment 11
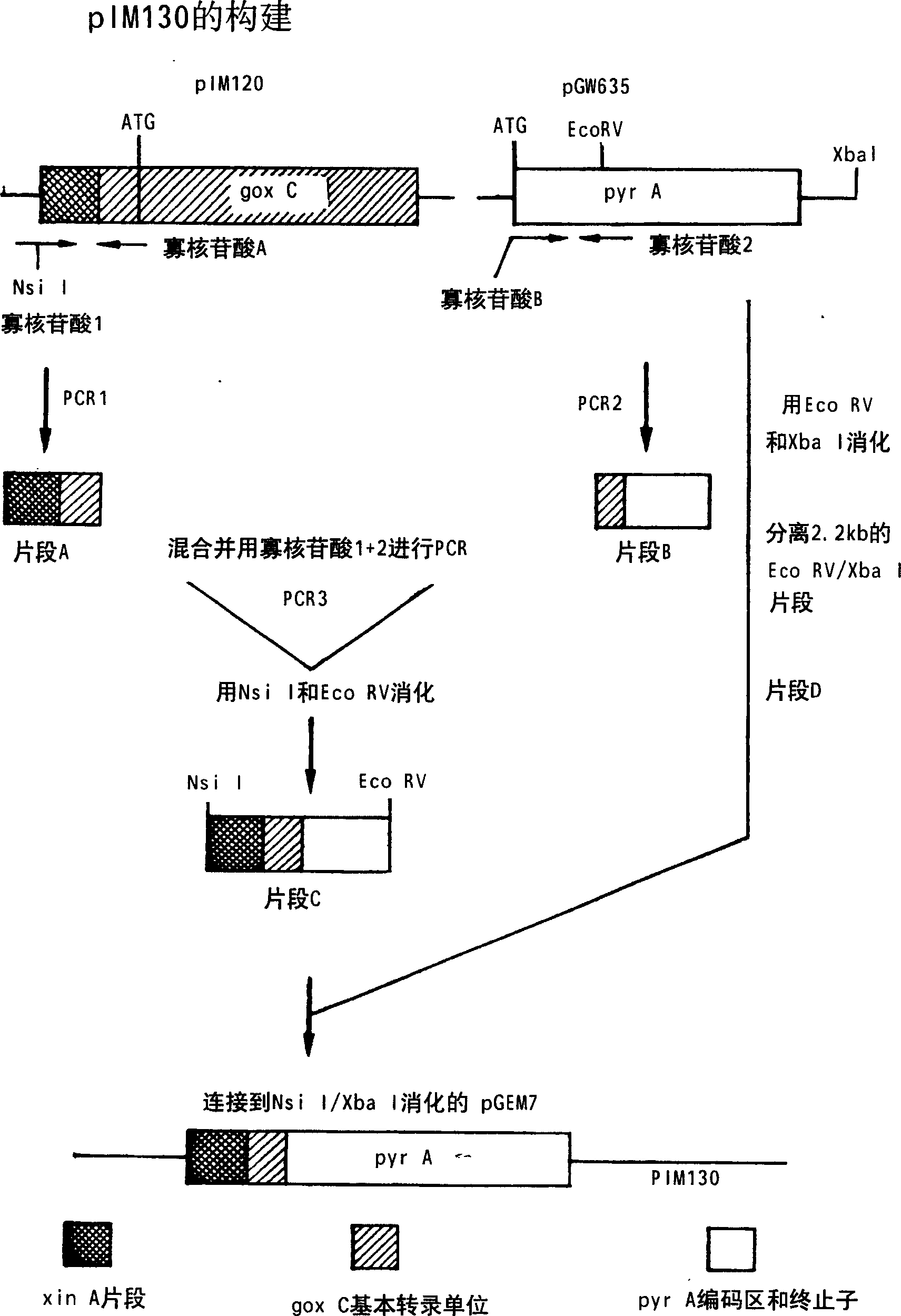
[0121] Example 1.1: Construction of selection plasmid pIM130
[0122] Such as figure 1 Plasmid pIM130 was selected for the construction shown in PCR1 with oligonucleotide 1 (SEQ ID NO: 1) 5'-CACAATGCATCCCCCTTTATCCGCCTGCCGT-3' (Formula 1) and oligonucleotide A (SEQ ID NO: 2) 5'-CAATTGCGACTTGGAGGACATGATGGGCAGATGAGGG -3' (Formula 2) generates a fragment from pIM120 (de Graaff et al., 1994). Oligonucleotide 1 was derived from positions 600-619 (SEQ ID NO: 5) of the Aspergillus tubingensis xlnA promoter (de Graaff et al., 1994) to which 10 nucleotides containing an NsiI site were added. The 3' end of oligonucleotide A was derived from the Aspergillus niger goxC transcription unit (Whittington et al., 1990) (positions 708-723) (SEQ ID NO: 6) ending before the translation initiation site, while its 5' end Derived from the coding region of the A. niger pyrA gene from the translation initiation site (Wilson et al., 1988) (positions 341-359, SEQ ID NO: 7).
[0123] By containing 10μl...
Embodiment 12
[0132] Example 1.2: Construction of plasmid pIM135
[0133] A second plasmid containing the goxC basic transcription unit fused to the pyrA coding and termination regions was constructed from plasmid pIM120. A fragment was generated from plasmid pIM120 in PCR4 using oligonucleotide 3 and oligonucleotide 2 (Formula 3) whose sequence was 5'-CACAATGCATCGTATAAGTAACCTCGTTCG-3' (Formula 5) derived from From the goxC basic transcription unit (positions 640-660, SEQ ID NO: 6), and 10 nucleotides containing an NsiI site were added. The resulting fragment was isolated from the gel as described in Example 1.1, digested with NsiI and EcoRV and cloned together with the 2.2 kb EcoRV / XbaI fragment of pGW635 into the XbaI / NsiI digested plasmid pGEM-7Zf(+) to give the plasmid pIM135.
[0134] Plasmid pIM135 can be used as a construction vector to prepare the UAS vector carrying any desired inducible enhancer sequence or activator sequence, genes involved in metabolism of the present inventio...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com