Chris-like virus, compositions, vaccines and uses thereof
a technology of chris-like virus and composition, applied in the field of chris-like virus, compositions, vaccines, can solve the problems of complex replication of all negative-strand rna viruses, and alone cannot synthesize the required rna-dependent rna polymeras
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
examples
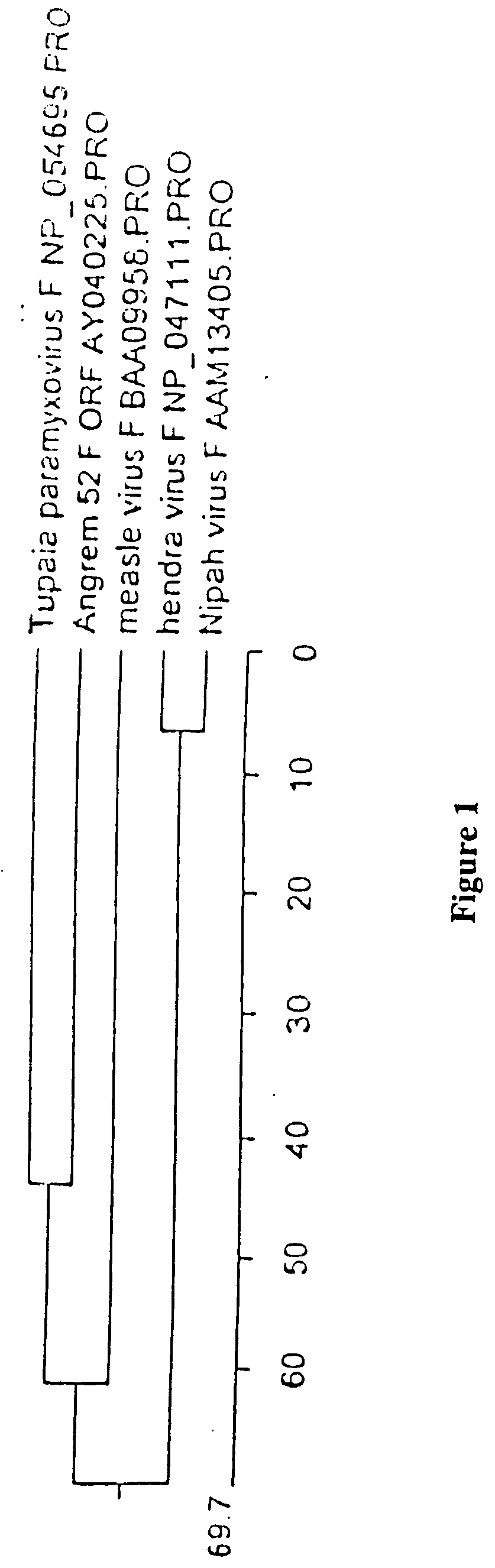
[0162] Sequences encoding genes of a new and novel paramyxovirus, Chris-like virus (CV) have been identified. Nucleotide sequences identified by Liang et al (Liang, X. et al (2003) J Am Soc Nephrol 14:1443-1451; Liang, X. et al (2003) Hypertens Res. 26(3):225-235) as novel human genes upregulated by angiotensin II (Ang II) in human kidney glomerular mesangial cells are actually RNA from a novel virus. The Liang et al nucleotide sequences are available from the National Center for Biotechnology Information (NCBI) (ncbi.nlm.nih.gov) as accession numbers AF367870, AY040225 and AY032980 (SEQ ID NOS: 1, 3 and 5) (Genbank entry AY032980 was submitted to the database earlier than and is a partial sequence which is encompassed entirely by AY040225, which contains additional 5′ end RNA sequence. Deduced amino acid sequences for AF367870, AY040225 and AY032980 are shown in SEQ ID NOS: 2, 4 and 6 respectively. However, there are several sequence discrepancies within the region of overlap. AY04...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Fraction | aaaaa | aaaaa |
| Composition | aaaaa | aaaaa |
| Nucleic acid sequence | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com