Use of mass labeled probes to detect target nucleic acids using mass spectrometry
a mass spectrometry and target nucleic acid technology, applied in combinational chemistry, biochemistry apparatus and processes, chemical libraries, etc., can solve the problems of limiting the number of labels that can be used simultaneously in a fluorescence detection scheme, multiplexed assays require more than just multiple tags, and detection apparatuses are difficult to achiev
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0053]This invention describes reagents, methods and kits that exploit circularising probes to characterise nucleic acids by mass spectrometry.
Definitions
[0054]The term ‘MS / MS’ in the context of mass spectrometers refers to mass spectrometers capable of selecting ions, subjecting selected ions to Collision Induced Dissociation (CID) and subjecting the fragment ions to further analysis.
[0055]The term ‘serial instrument’ refers to mass spectrometers capable of MS / MS in which mass analysers are organised in series and each step of the MS / MS process is performed one after the other in linked mass analysers. Typical serial instruments include triple quadrupole mass spectrometers, tandem sector instruments and quadrupole time of flight mass spectrometers.
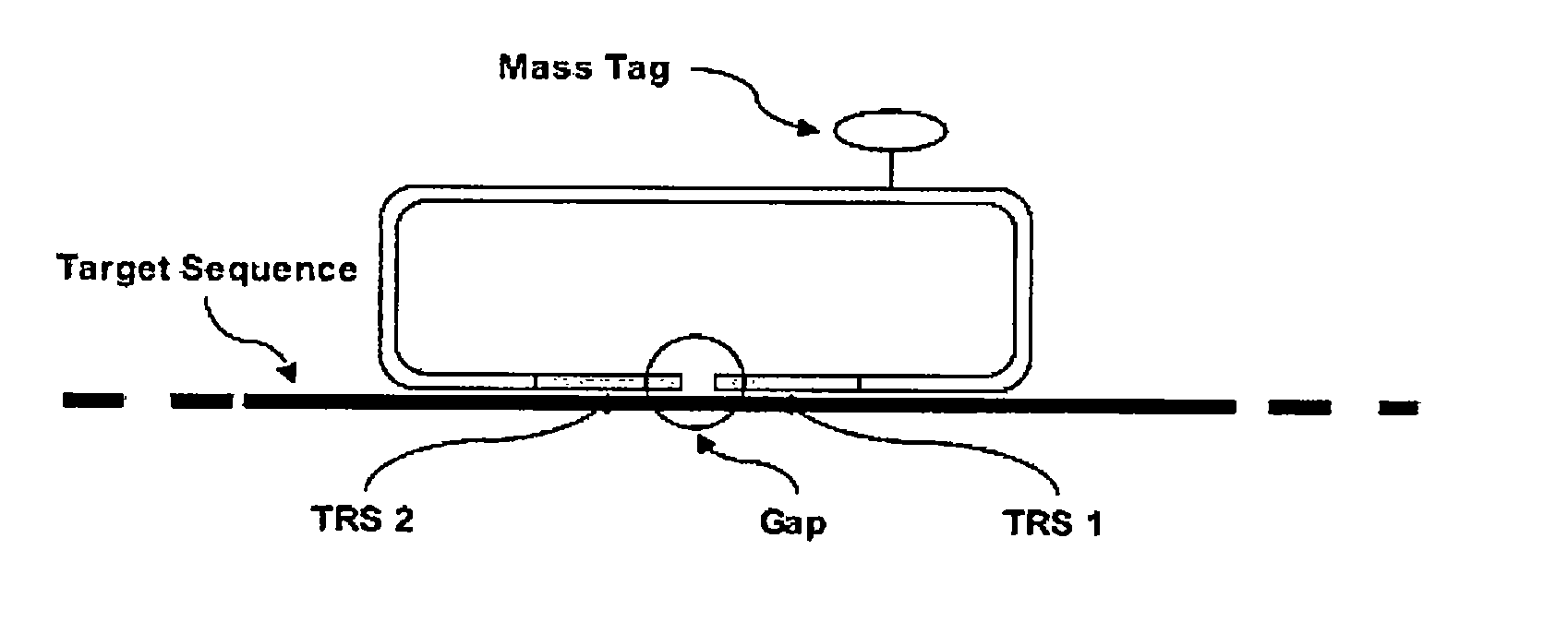
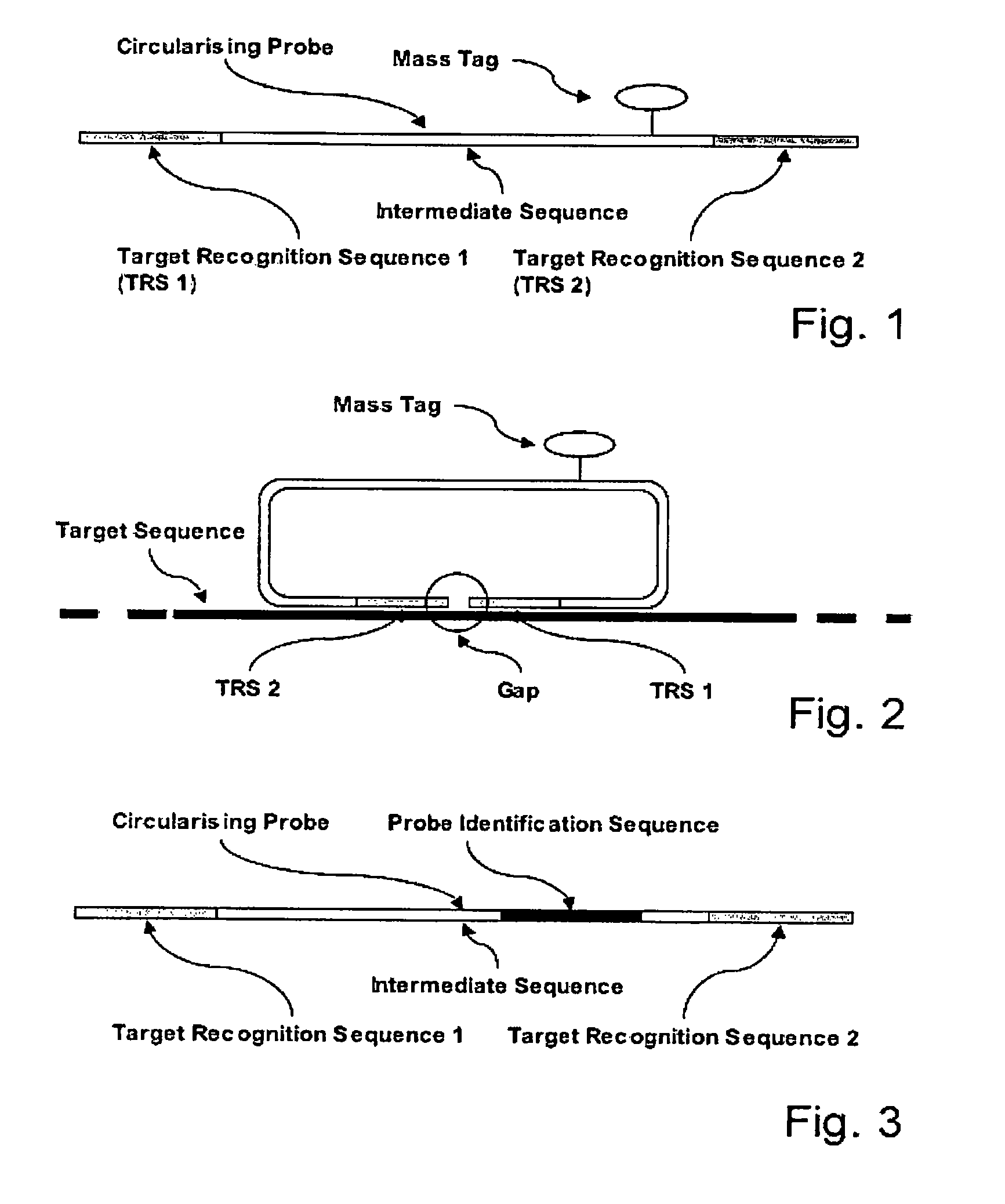
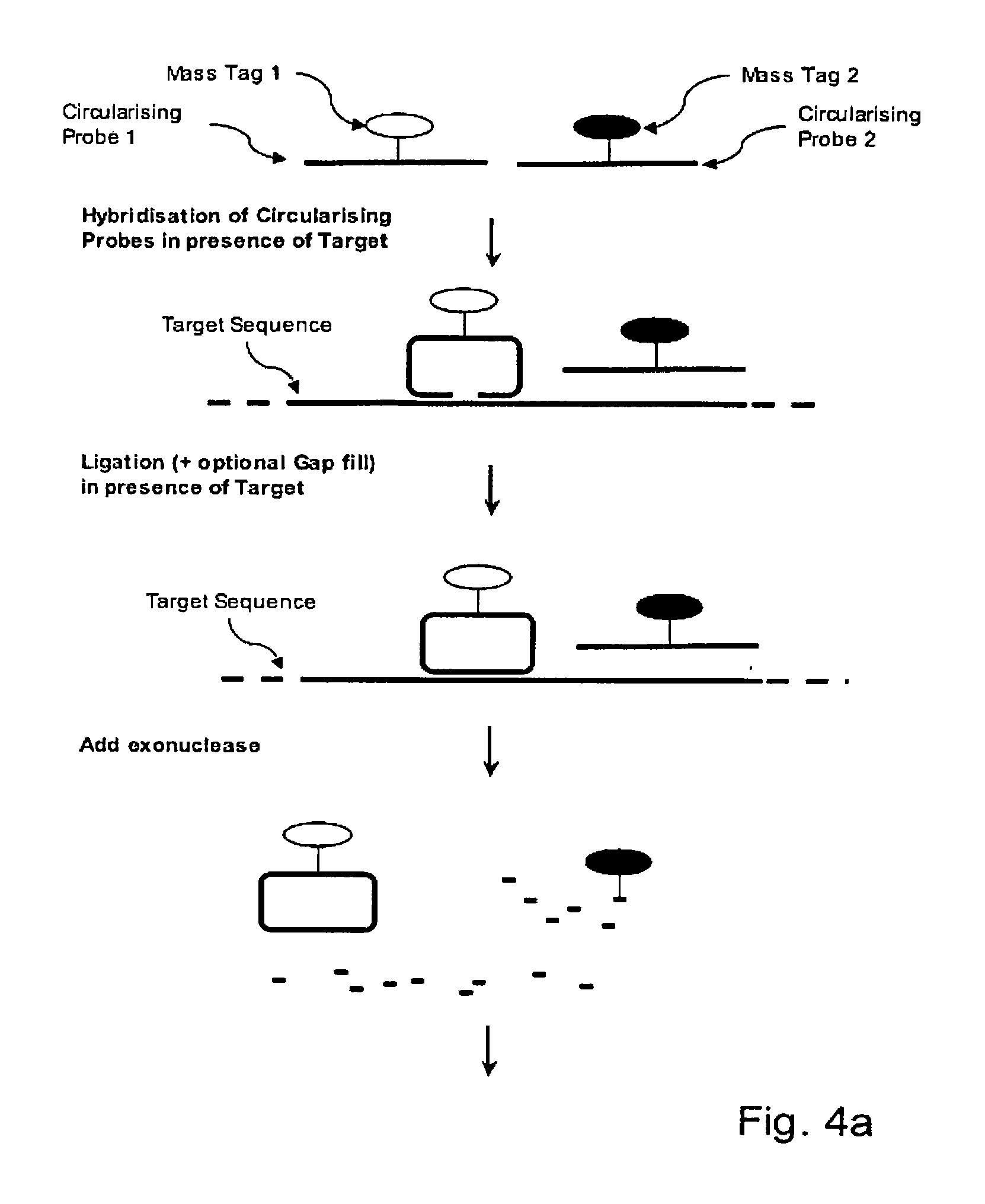
[0056]A Linear Circularising Probe (LCP) is probe sequence where the two termini of the probe comprise Target Recognition Sequences (TRS) that are designed to hybridise in juxtaposition on a target nucleic acid. The 3′ terminus of the pro...
PUM
| Property | Measurement | Unit |
|---|---|---|
| mass spectrometry | aaaaa | aaaaa |
| mass | aaaaa | aaaaa |
| affinity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com