DNA polymerase
a polymerase and dna technology, applied in the field of dna polymerases, can solve the problems of limiting preventing the efficient pcr amplification of damaged dna templates, and restricting the use of unnatural or modified nucleotide bases and the applications they enabl
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
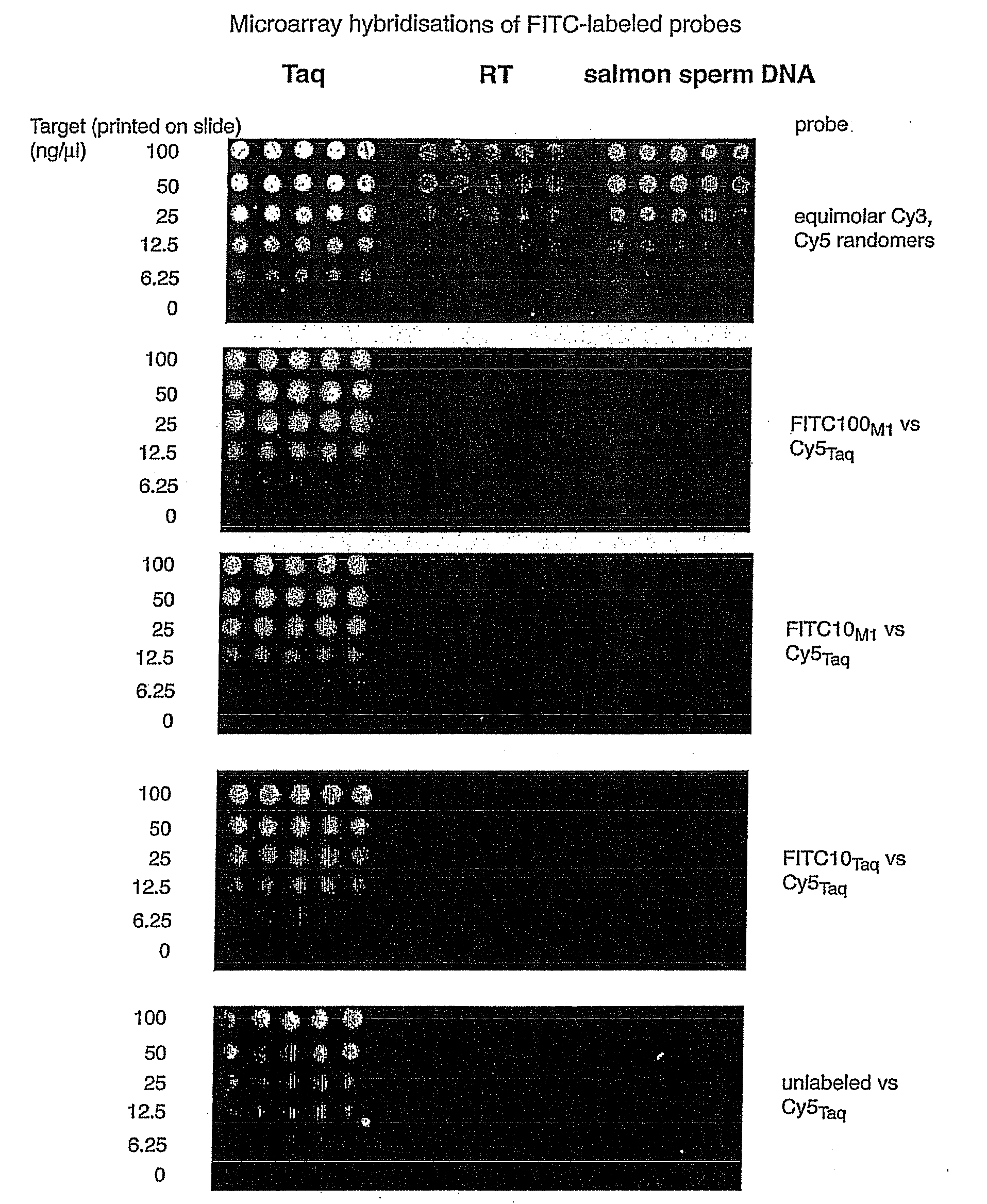
Image
Examples
example 1
General Methods
List of Primers:
[0173]
1:5′-CAG GAA ACA GCT ATG ACA AAA ATC TAG ATA ACG AGG GA-3′;(SEQ ID NO: 3)A•G mismatch 2:5′-GTA AAA CGA CGG CCA GTA CCA CCG AAC TGC GGG TGA CGC(SEQ ID NO: 4)CAA GCC-3′; C•C mismatch 3:5′-AAA AAT CTA GAT AAC GAG GGC AA-3′(SEQ ID NO: 13) 4:5′-ACC ACC GAA CTG CGG GTG ACG CCA AGC G-3′(SEQ ID NO: 14) 5:5′-GAA CTG CGG GTG ACG CCA AGC GCA 3′; A•A mismatch(SEQ ID NO: 15) 6:5′-CC GAA CTG CGG GTG ACG CCA AGC GG 3′; G•G mismatch(SEQ ID NO: 16) 7:5′-GAA CTG CGG GTG ACG CCA AGC GCG-3′; G•A mismatch(SEQ ID NO: 17) 8:5′-AAA AAT CTA GAT AAC GAG GGC AA-3′(SEQ ID NO: 18) 9:5′-CCG ACT GGC CAA GAT TAG AGA GTA TGG-3′(SEQ ID NO: 19)10:5′-GAT TTC CAC GGA TAA GAC TCC GCA TCC-3′(SEQ ID NO: 20)11:5′-GGC AGA CGA TGA TGC AGA TAA CCA GAG C-3′(SEQ ID NO: 21)12:5′-GCC GAT AGA TAG CCA CGG ACT TCG TAG-3′(SEQ ID NO: 22)13:5′-GGA GTA GAT GCT TGC TTT TCT GAG CC-3′(SEQ ID NO: 23)14:5′-GAG TTC GTG CTT ACC GCA GAA TGC AG-3′(SEQ ID NO: 24)15:5′-ACC GAA CTG CGG GTG ACG CCA AGC G 3′(SEQ ...
example 2
[0182]Kinetic analysis. Extension and incorporation kinetics of M1 and M4 for a selection of mismatches were measured using a gel-based steady-state kinetic assay (Goodman) (Tables 1 & 2). M1 and M4 respectively extend a C·C mispair 390 and 75-fold more efficiently than wtTaq. Examination of the other most disfavored mismatches (G·A, A·G, A·A, G·G) reveals generic, although less pronounced, increases of extension efficiencies, as suggested by the PCR assay (FIG. 4, FIG. 5). The gain in extension efficiency derives predominantly from increased relative Vmax values for the mutant polymerases, while Km for nucleotide substrates remains largely unchanged. For most DNA polymerases the relative efficiency of extending a given mispair (f0ext) is similar to the relative efficiency of forming it (finc) (Goodman 1993, Goodman 1990, Washington 2001). Indeed, M1 and M4 respectively incorporate dCTP opposite template base C 206- and 29-fold more efficiently than wtTaq (Table 2).
TABLE 2Steady-sta...
example 3
[0183]Translesion synthesis. Transversion mispairs represent distorting deviations from the cognate duplex structure. We therefore investigated if M1 and M4 were capable of processing other deviations of the DNA structure such as lesions in the template strand. Using a gel-extension assay we investigated their ability to traverse an abasic site and a cis-syn thymine pyrimidine dimer (CPD) template strand lesion. In control assays using an undamaged template, wtTaq, M1 and M4 efficiently and rapidly extended primers to the end of the template (FIG. 5). On the template containing an abasic site, wtTaq efficiently inserts a base opposite the lesion but, further extension is largely abolished. In contrast, both M1 and M4 are able to extend past the lesion and to the end of the template. The size of the final product is similar to that observed on the undamaged template indicating that bypass occurred without deletions. Perhaps the most striking example of the proficiency of M1 and M4 to...
PUM
| Property | Measurement | Unit |
|---|---|---|
| volume | aaaaa | aaaaa |
| diameter | aaaaa | aaaaa |
| volume | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com