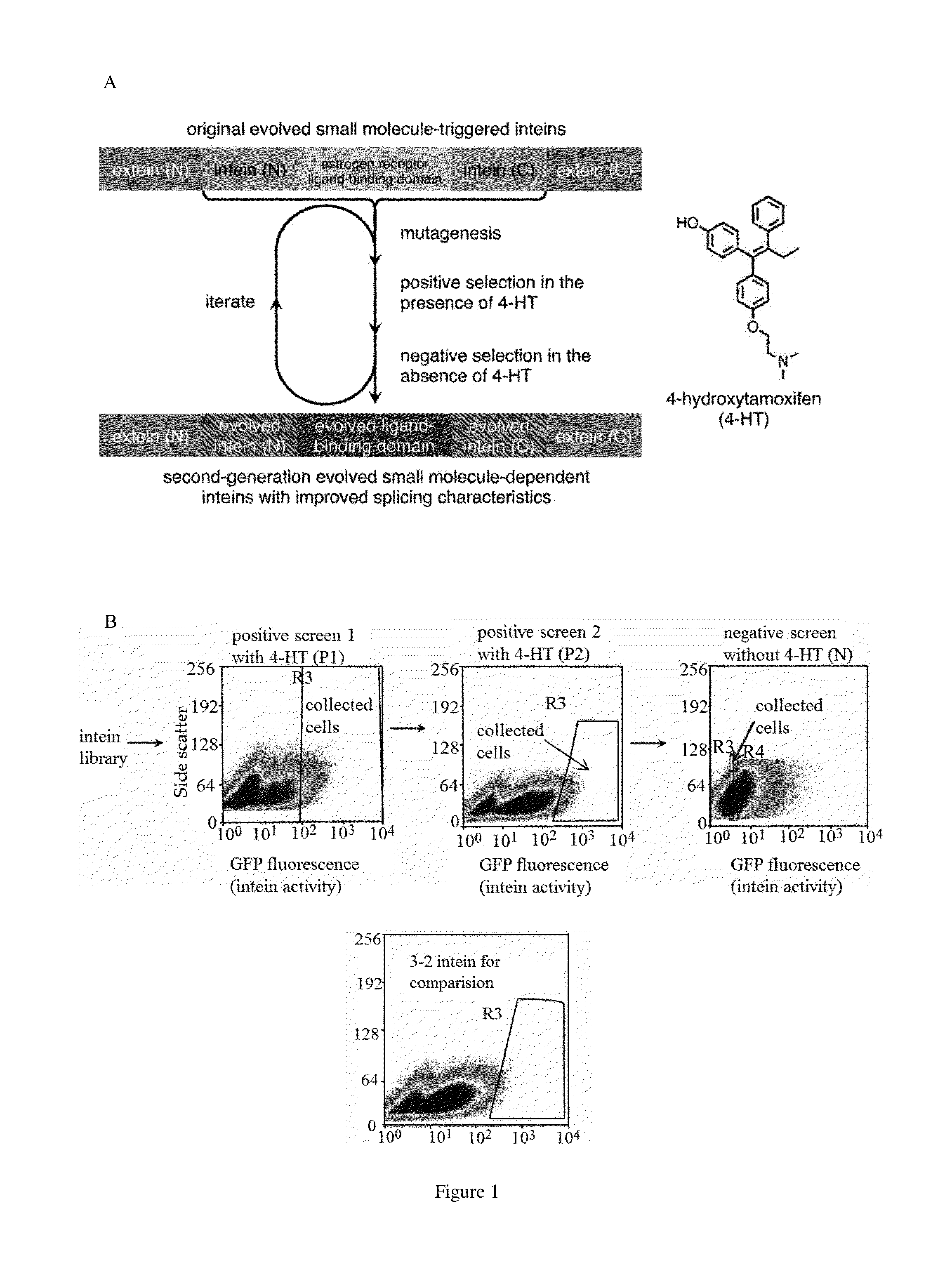
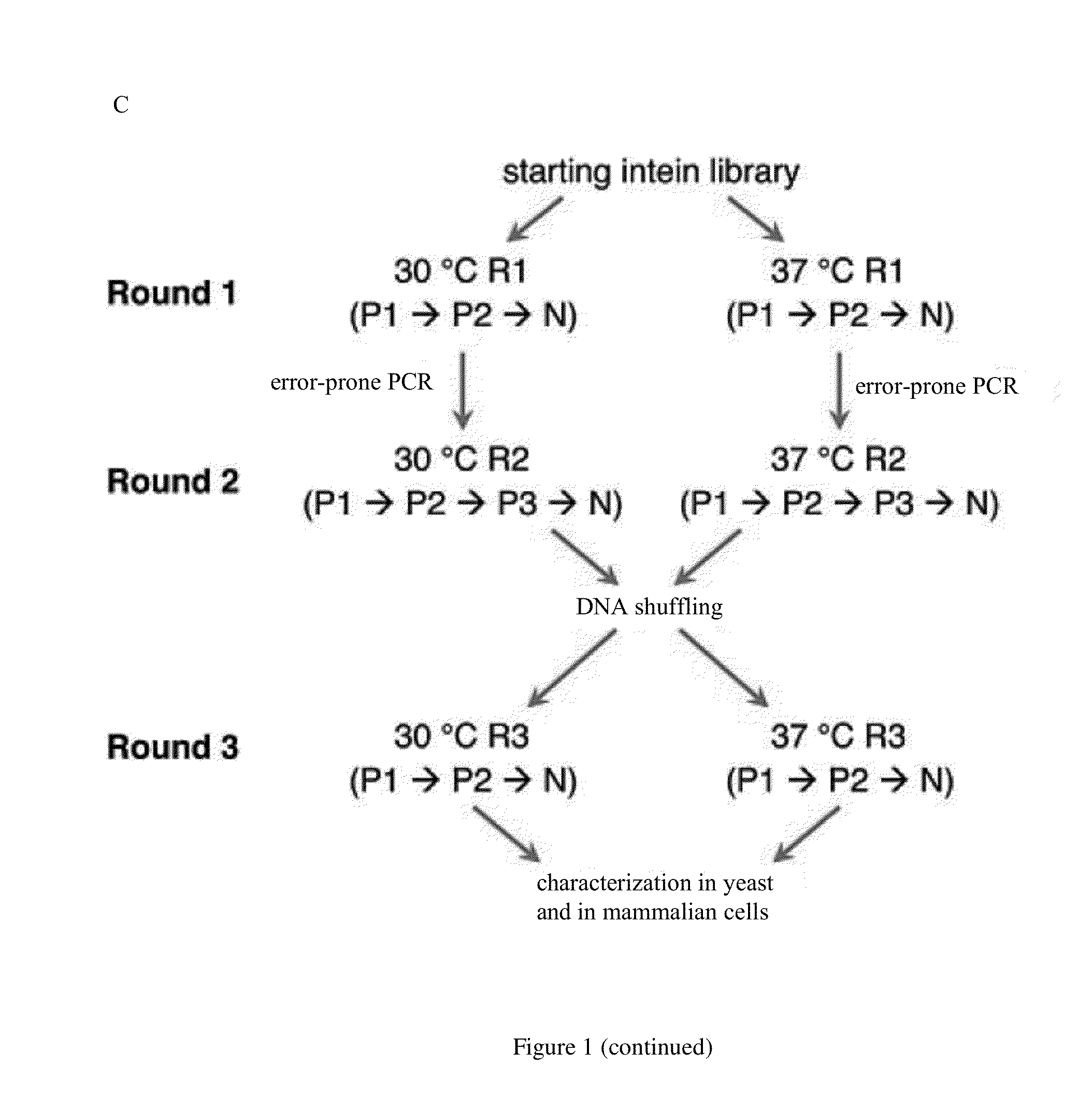
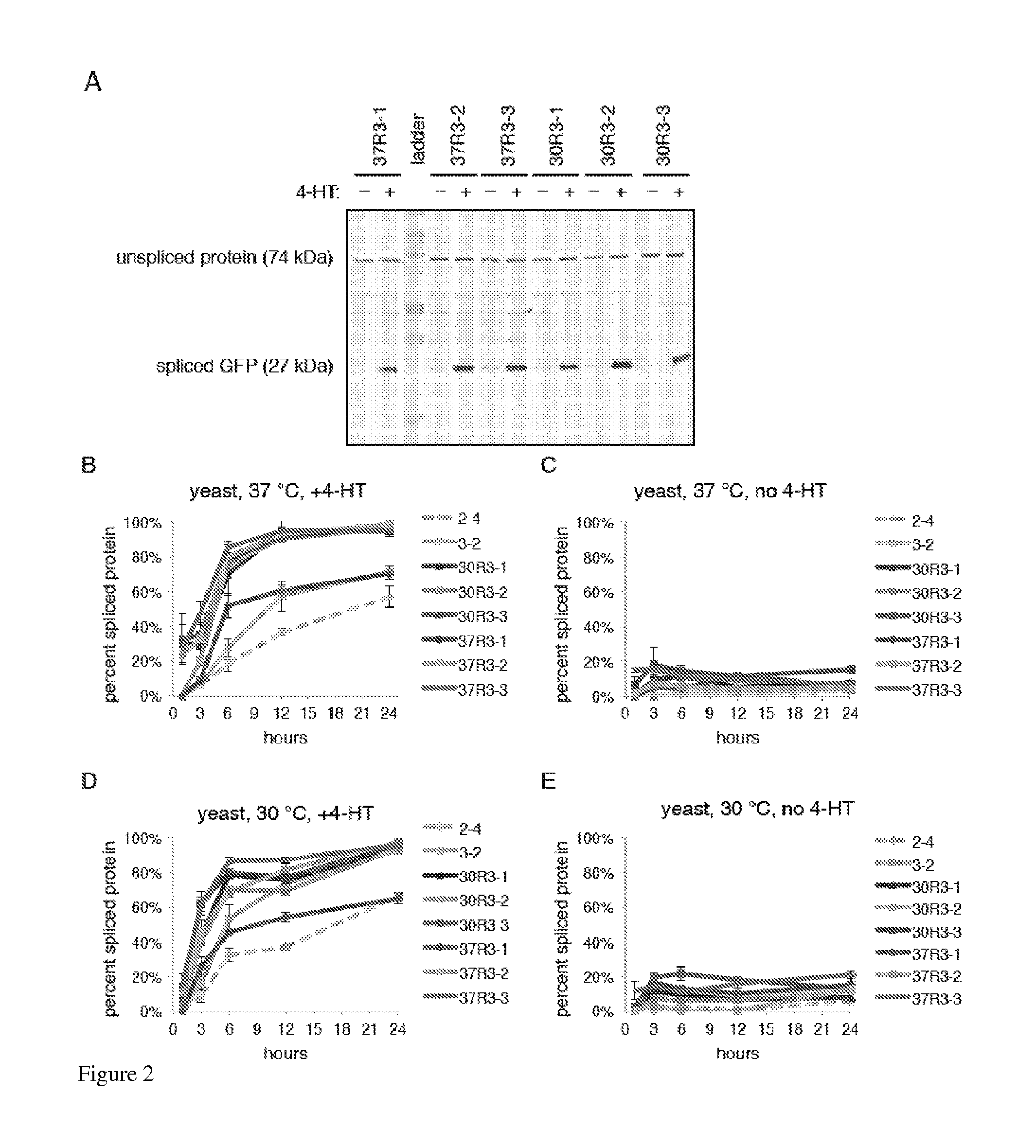
Small molecule-dependent inteins and uses thereof
a small molecule, intein technology, applied in the direction of peptide sources, specific cell targeting fusions, peptides, etc., can solve the problems of no natural inteins, no natural inteins, regulated by small molecules, conventional ligand-dependent inteins, etc., to achieve lower background splicing, slow splicing, and low splicing efficiencies
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
examples
Materials and Methods
Yeast Strains and Media
[0090]Media consisted of yeast nitrogen base (Sigma), 4% dextrose, and synthetic drop out supplements lacking uracil (MP Biomedical). Yeast were cultured in liquid medium or on agar plates at 30° C. The yeast strain RDY98 (Erg6del::TRP1 pdr1del::KanMX pdr3::HIS3 ade2-1 trp1-1 his3-11,15 ura3-52 leu2-3,112 can1-100) was provided by Professor Allen Buskirk at Brigham Young University. Protein induction was performed in media consisting of yeast nitrogen base (Sigma), 4% galactose, 4% raffinose, 0.4% dextrose, synthetic drop out supplements lacking uracil (MP Biomedical), and 1% of 100× penicillin-streptomycin solution (Cellgro) at 30 ° C.
Mammalian Cell Culture
[0091]HEK293 cells were cultured in Dulbecco's modified Eagle medium (DMEM):F12 medium with 10% fetal bovine serum (FBS) and 1% of 100× penicillin-streptomycin solution (Cellgro) according to standard protocols. Transient transfections were performed using Effectene (Qiagen) following t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com