Diagnostic and therapeutic methods for cancer
a technology of pdl1 and axis binding antagonist, applied in the direction of instruments, drug compositions, peptide/protein ingredients, etc., can solve the problems of difficult timely detection and treatment, and achieve the effect of responsiveness to treatment and responsiveness to treatmen
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
on Between Immune-Score Expression Levels of (i) PD-L1, CXCL9, and IFNG or (ii) PD-L1, IFNG, GZMB, and CD8A and Clinical Response of Patients Having Non-Small Cell Lung Cancer (NSCLC) to Treatment with Atezolizumab (MPDL3280A)
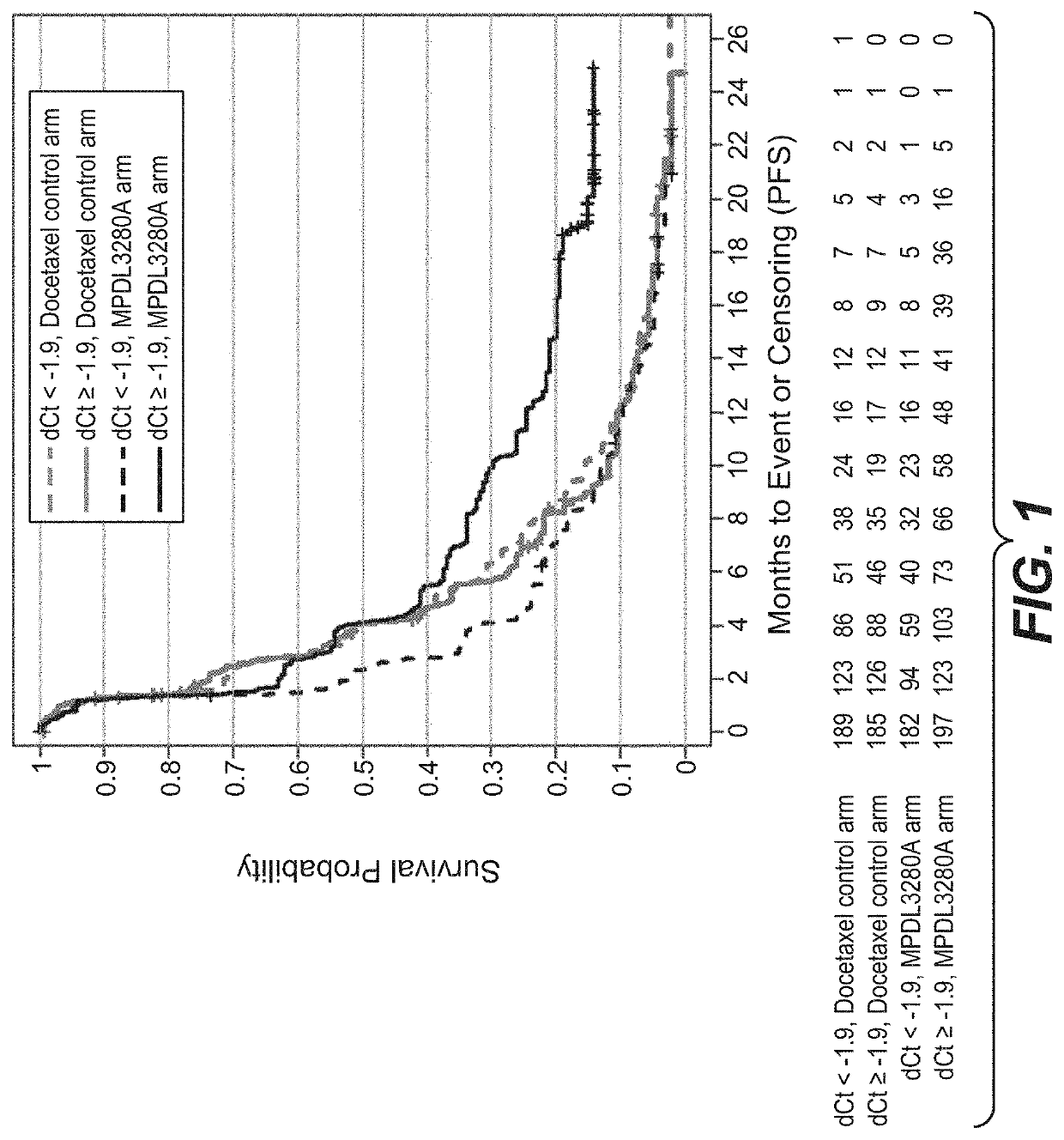
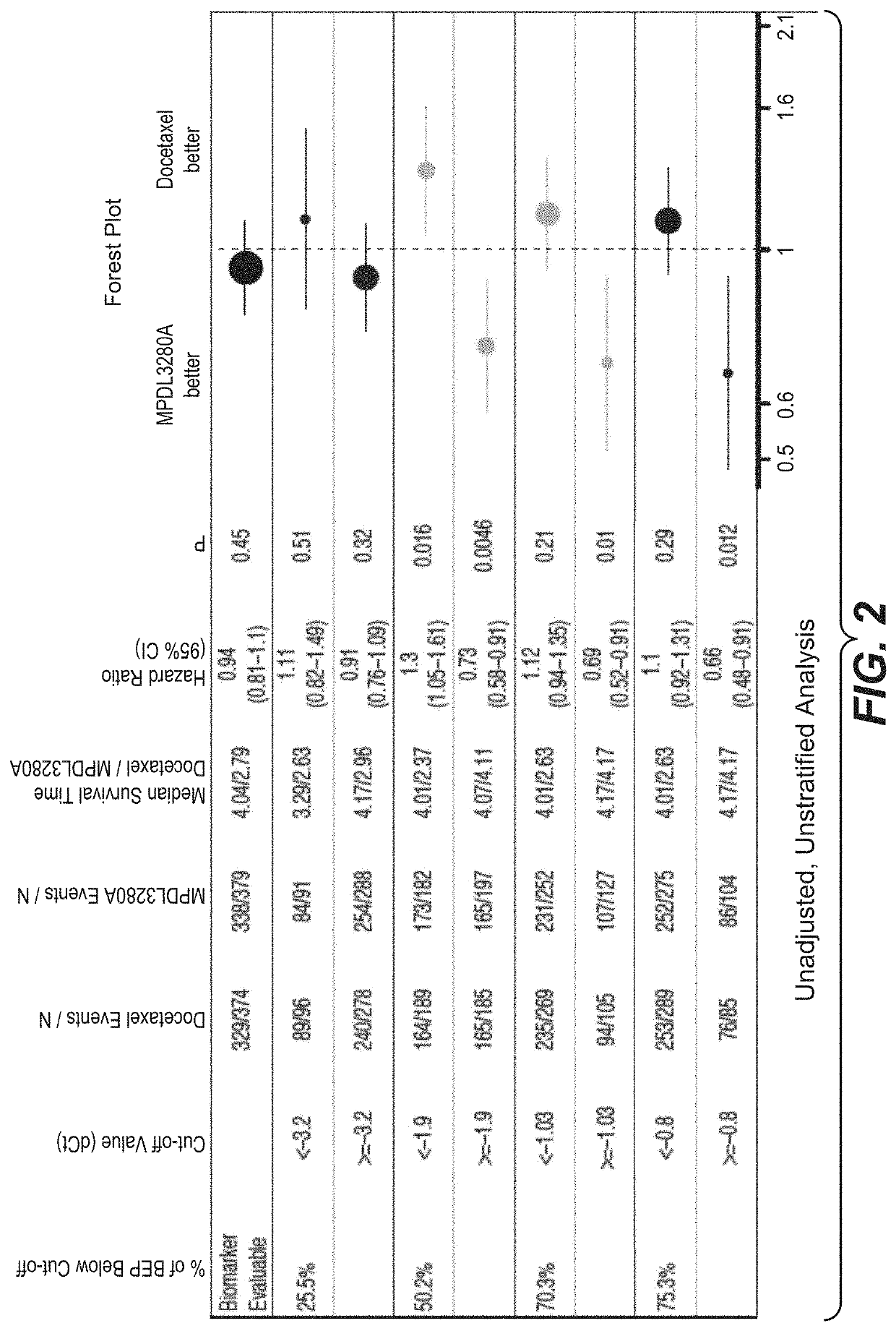
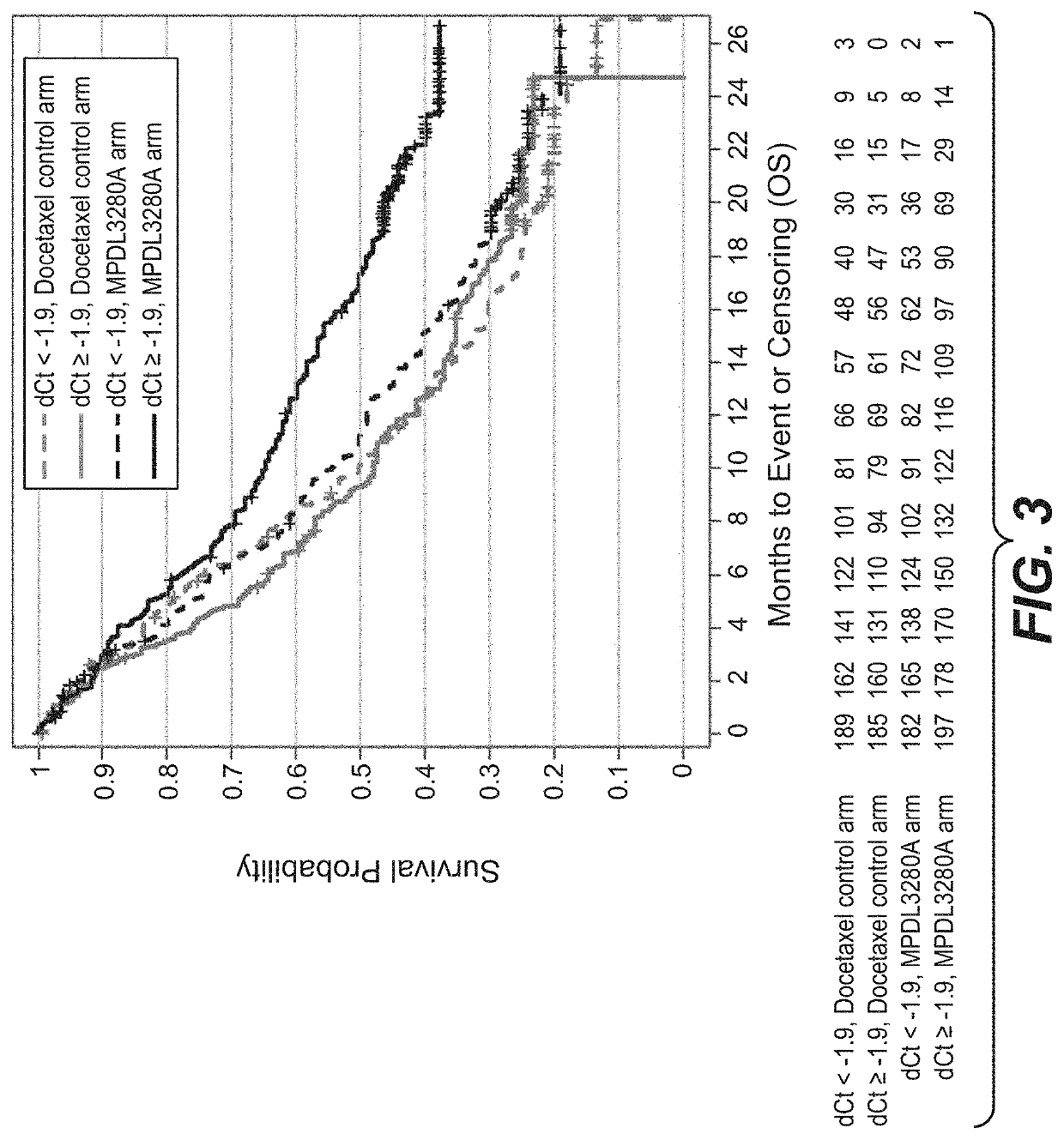
[0765]An RNA-based molecular assay was used to evaluate the association between clinical response to treatment with atezolizumab (MPDL3280A), an anti-PD-L1-antibody, and immune-score expression levels of (i) PD-L1, CXCL9, and IFNG or (ii) PD-L1, IFNG, GZMB, and CD8A in patients with non-small cell lung cancer (NSCLC) enrolled in a phase III clinical trial in which atezolizumab was administered as a monotherapy.
Study Design
[0766]The OAK (Clinical Trial ID No.: NCT02008227) patient population evaluated for (i) PD-L1, CXCL9, and IFNG and (ii) PD-L1, IFNG, GZMB, and CD8A expression levels consisted of 753 patients. Patients were eligible for enrollment in the OAK Trial if they had locally advanced or metastatic (e.g., stage IIIB, stage IV, or recurrent) NSCLC; dise...
example 2
on Between Expression Levels of PD-L1, CXCL9, and IFNG and Clinical Response of Patients Having NSCLC to Treatment with Atezolizumab (MPDL3280A)
[0778]An RNA-based molecular assay was used to evaluate the association between clinical response to treatment with atezolizumab (MPDL3280A), an anti-PD-L1 antibody, and expression levels of PD-L1, CXCL9, and IFNG in individuals with NSCLC enrolled in a phase II clinical trial in which atezolizumab was administered as a monotherapy.
Study Design
[0779]The POPLAR (Clinical Trial ID No.: NCT01903993) patient population evaluated for PD-L1, CXCL9, and IFNG expression levels consisted of 215 patients. Patients were eligible for enrollment in the POPLAR study if they had locally advanced or metastatic (e.g., stage IIIB, stage IV, or recurrent) NSCLC; disease progression during or following treatment with a prior platinum-containing regimen for locally advanced, unresectable / inoperable, or metastatic NSCLC, or disease recurrence within 6 months of t...
example 3
on Between Expression Level of PD-L1, CXCL9, and IFNG and Clinical Response of Patients Having UBC to Treatment with Atezolizumab (MPDL3280A)
[0785]An RNA-based molecular assay was used to evaluate the association between clinical response to treatment with atezolizumab (MPDL3280A), an anti-PD-L1 antibody, and expression levels of PD-L1, CXCL9, and IFNG in individuals with advanced urothelial bladder cancer (UBC) enrolled in a phase II clinical trial (the IMvigor210 Trial) in which atezolizumab was administered as a monotherapy.
Study Design
[0786]Pre-treatment tumor specimens from patients with advanced UBC who were in Cohort 2 of the Phase II IMvigor210 Trial (Clinical Trial ID No.: NCT02108652) were evaluated for the expression level of PD-L1, CXCL9, and IFNG. Patients were eligible for enrollment in Cohort 2 of the IMvigor210 Trial if they had histologically or cytologically documented locally advanced or metastatic transitional cell carcinoma or the urothelium (e.g., renal pelvis,...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Fraction | aaaaa | aaaaa |
| Fraction | aaaaa | aaaaa |
| Fraction | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com