Biomarkers predictive of therapeutic responsiveness to chimeric antigen receptor therapy and uses thereof
a chimeric antigen receptor and biomarker technology, applied in the field of cancer biomarkers, can solve the problems of difficult to achieve clinical effectiveness, difficult to detect, and difficult to detect, and achieve the goal of clinical efficacy, so as to and reduce the risk of subject relapse
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
ation of Novel Transcriptional Gene Signatures that Predict Subject Response to CD19 CAR-Expressing Cell Therapy in Chronic Lymphoid Leukemia (CLL) and Acute Lymphoblastic Leukemia (ALL) Using Whole Genome RNAseq and Unbiased Feature Selection
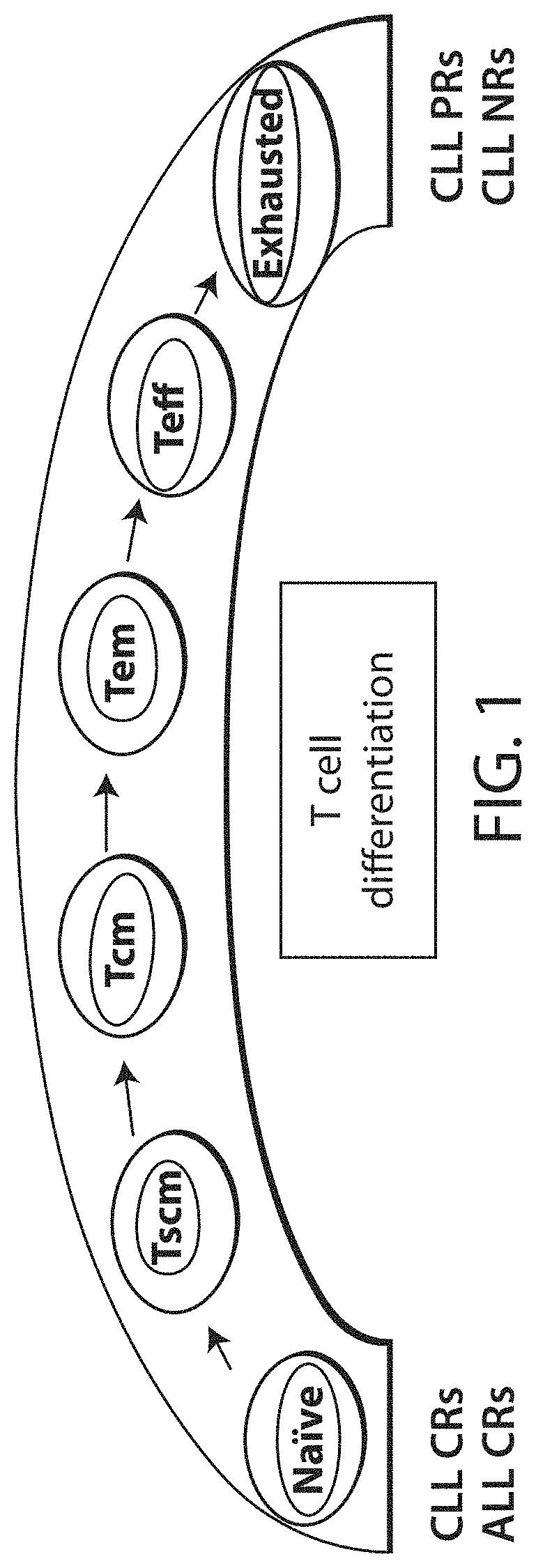
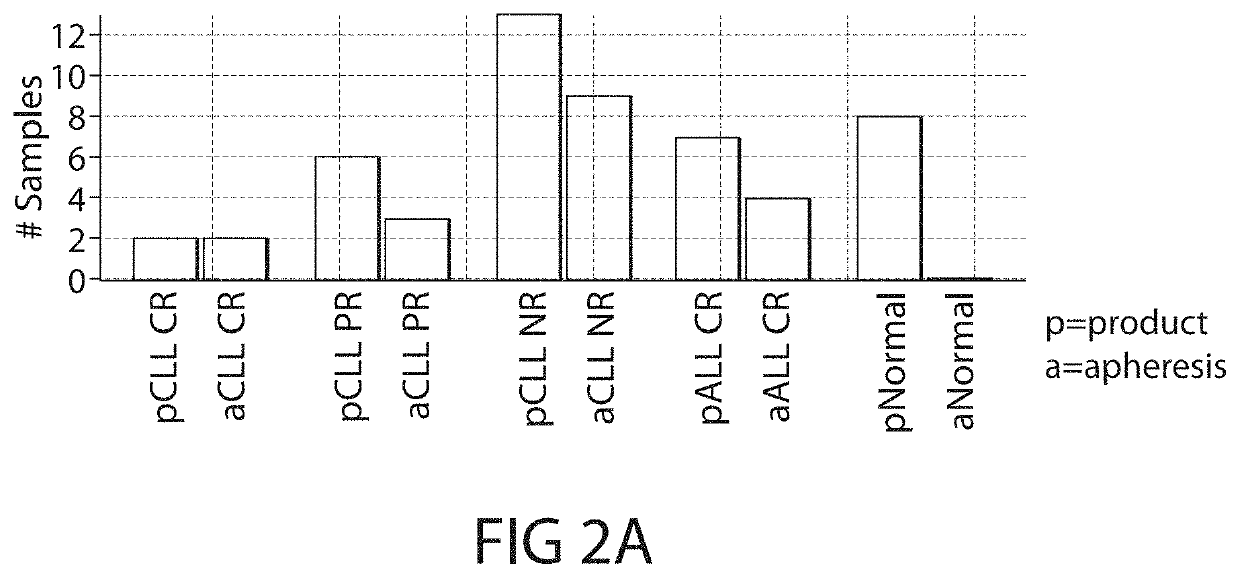
[0917]The present Example describes the identification of novel transcriptional gene signatures that predict patient response to CD19 CAR-expressing cell (e.g., T cell, NK cell) therapy (e.g., CTL019 therapy) in Chronic Lymphoid Leukemia (CLL) and Acute Lymphoblastic Leukemia (ALL), for use in accordance with the present invention.
[0918]Among other things, the present Example describes novel gene signatures based on mRNA expression levels of selected genes in apheresis and manufactured CD19 CAR-expressing cell (e.g., T cell, NK cell) product samples (e.g., CTL019) prior to re-infusion.
[0919]In particular, the present Example describes methods of unbiased feature selection to discover novel gene signatures that predict patient response to CD19 C...
example 2
ation of Novel Transcriptional Gene Signatures which Predict Subject Response to CD19 CAR-Expressing Cell Therapy in Chronic Lymphoid Leukemia (CLL) and Acute Lymphoblastic Leukemia (ALL) Using Gene Set Analysis and Differential Expression Analysis
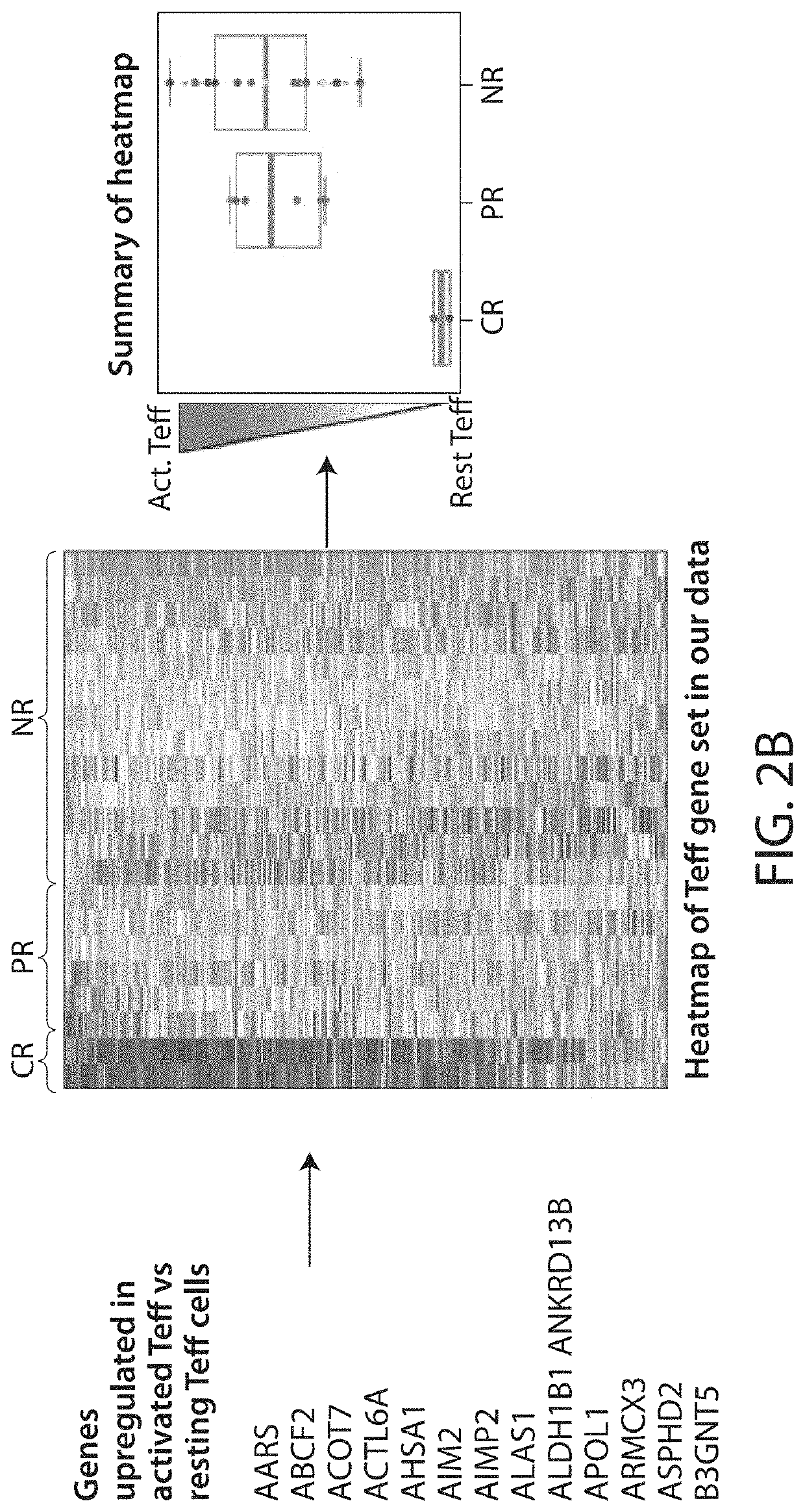
[0926]The present Example describes the identification of novel transcriptional gene signatures that predict patient response to CD19 CAR-expressing cell (e.g., T cell, NK cell) therapy (e.g., CTL019) in CLL and ALL, for use in accordance with the present invention.
[0927]In particular, the present Example describes methods of Gene Set Analysis to discover novel gene signatures, for use in accordance with the present invention.
[0928]Among other things, the present Example describes novel gene signatures based on Gene Set Analysis, that are predictive of patent response to CD19 CAR-expressing cell (e.g., T cell, NK cell) therapy (e.g., CTL019). Gene set analysis was performed on gene sets described in Example 1, and with gene sets from three...
example 3
c Flow Cytometry-Based Assays
[0973]Prognostic flow cytometry-based assays are developed to screen subjects with cancer (e.g., patients with a hematological cancer such as ALL and CLL) for CAR-expressing cell (e.g., T cell, NK cell) therapy, e.g., CD19 CAR-expressing cell therapy as described herein such as, e.g., CTL019 therapy. In some embodiments, subjects are participating in clinical trials.
[0974]A sample (e.g., a blood sample) is isolated from a patient and a fluorescent flow cytometry-based assay is performed screening for one or more cell surface or secreted biomarkers described in Examples 1 and 2. An exemplary list of markers that are measured, e.g., by flow cytometry if cell surface-expressed, or by ELISA if secreted, and whose expression values predict patient response to CAR-expressing cell (e.g., T cell, NK cell) therapy, e.g., CD19 CAR-expressing cell therapy as described herein such as, e.g., CTL019 therapy includes, but is not limited to, genes listed in Table 8.
TABL...
PUM
| Property | Measurement | Unit |
|---|---|---|
| time course | aaaaa | aaaaa |
| cell surface | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com