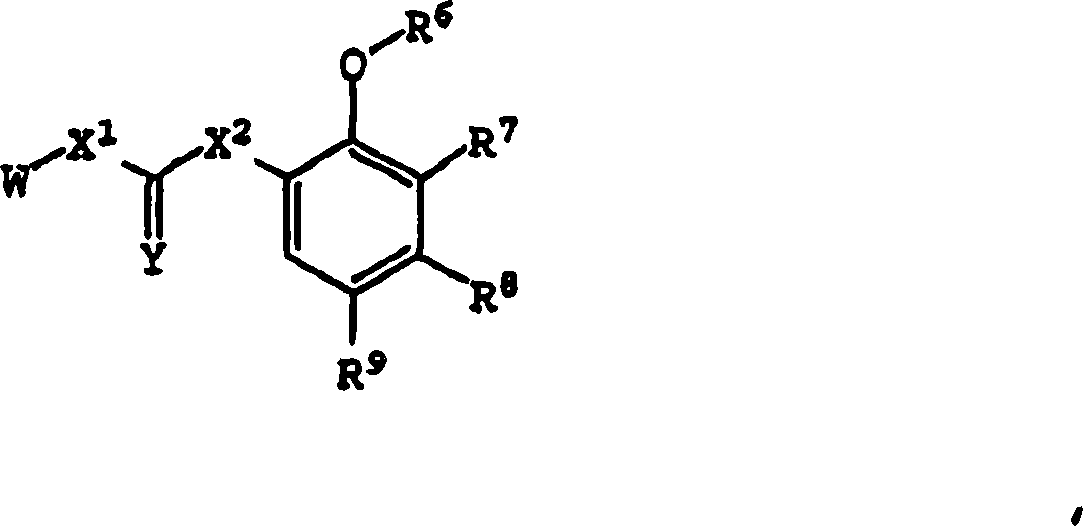
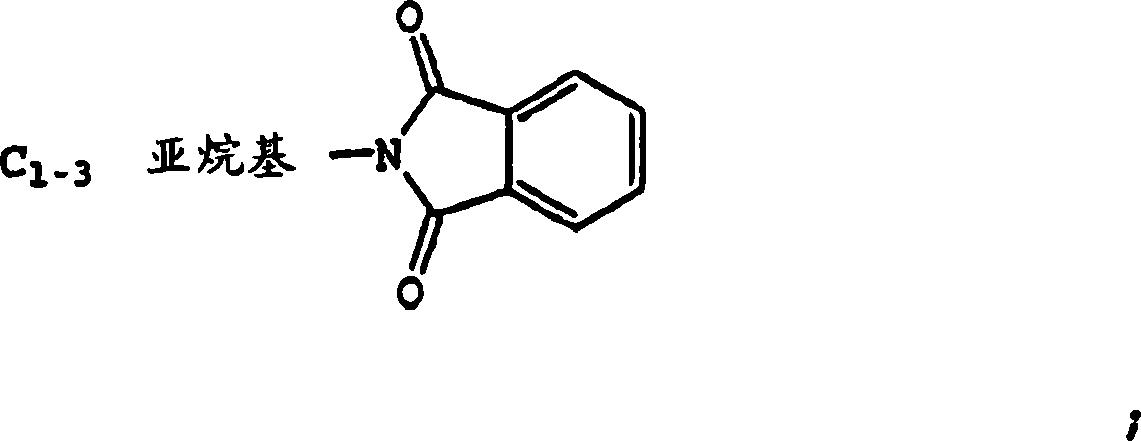
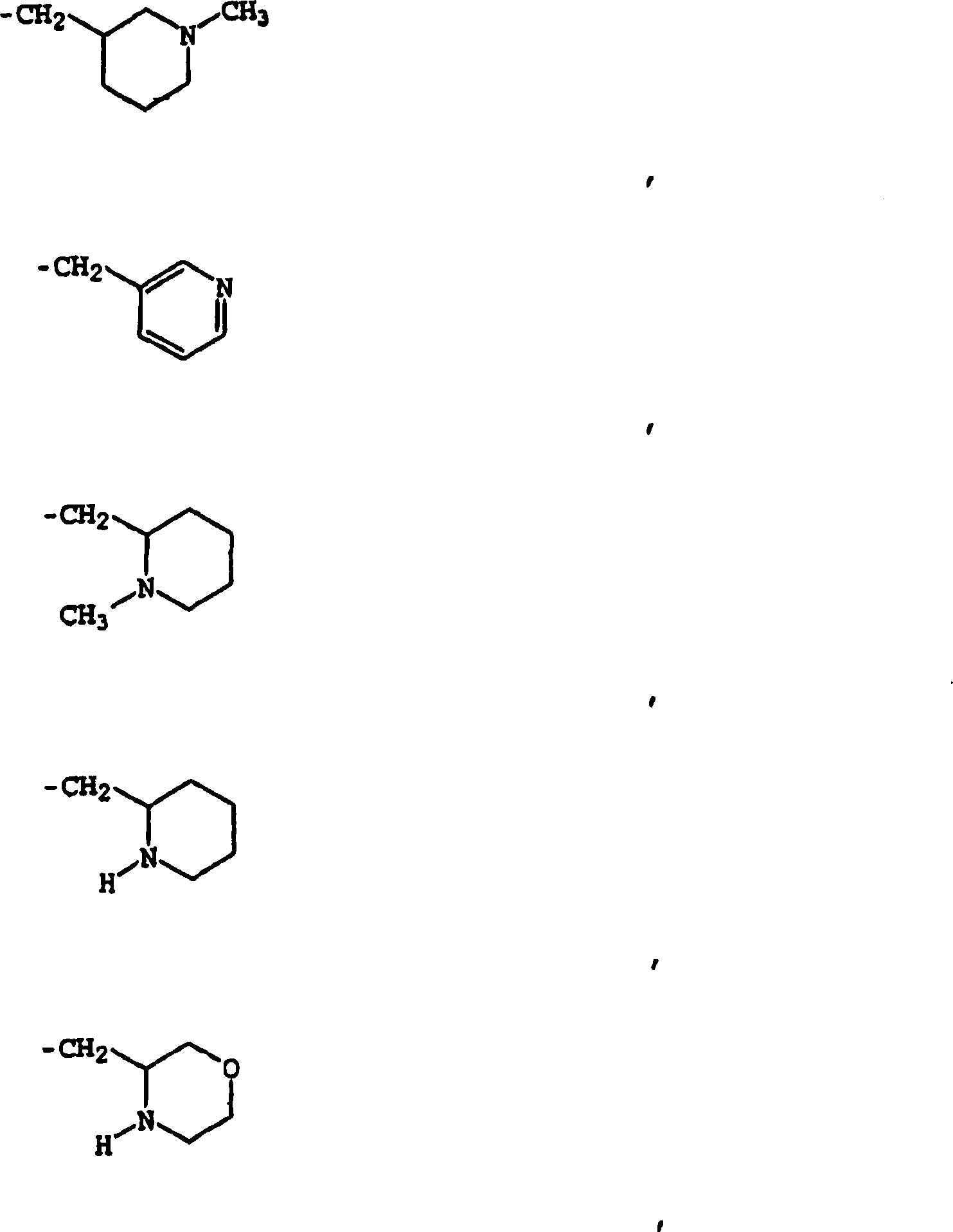
Bisarylurea derivatives useful for inhibiting CHK1
An aryl and heteroaryl technology, applied in the field of enzyme-inhibiting compounds, can solve the problems of increasing the sensitivity of the topoisomerase inhibitor BNP1350, exceeding the dose of caffeine, and not being a treatment option
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0351] Determination of ICs for Chk1 inhibitors 50 value
[0352] The cDNA for human Chk1 has been identified and cloned as previously described in International Application Publication No. WO 99 / 11795, filed September 4,1998. A FLAG_tag was inserted in frame at the amino terminus of full-length Chk1. The 5' primer contained an EcoRI site, a Kozak sequence, and also encoded FLAG for affinity purification using the M2 antibody (Sigma, Saint Louis, IL) _ Label. The 3' primer contains a SalI site. The PCR amplified fragment was cloned into pCI-Neo (Invitrogen, Carlsbad, CA) as an EcoRI-SalI fragment and then subcloned into pFastBacI (Gibco-BRL, Bethesda, MD) as an EcoRI-NotI fragment. Recombinant baculoviruses were prepared as described in the Gibco-BRL Bac-to-Bac manual and used to infect Sf-9 cells grown in CCM3 medium (HyClone Laboratories, Logan, UT) to express plus with FLAG _ Tagged Chk1 protein.
[0353] Purification of FLAG-added FLAG from frozen blocks of baculovi...
Embodiment 2
[0356] selectivity
[0357] Relative to one or more other protein kinases, i.e., DNA-PK, Cdc2, casein kinase I (CKI), Chk2, p38 MAP kinase, ERK kinase, protein kinase A (PKA), and / or calcium-calmodulin Protein Kinase II (CaM KII) tests the selectivity of the Chk1 inhibitors of the present invention. With the exception of Chk2, assay procedures for all of these kinases have been previously described in the literature, including US Patent Publication 2002-016521 Al and US Patent Application Serial No. 08 / 184,605, filed January 21, 1994, which are incorporated herein by reference.
[0358] The activity of the compounds on Chk2 was determined as follows: at room temperature, in 4 mM ATP, 1 mCi [ 32 P]γ-ATP, 20mM Hepes pH7.5, 5mM MgCl 2 128 ng of purified His-tagged Chk2 was incubated with up to 100 mM Chk1 inhibitor in the presence of 0.25% NP40 for 20 minutes. The reaction was stopped with phosphoric acid at a final concentration of 150 mM, and 5 / 8 of the reaction mixture wa...
Embodiment 3
[0361] Chk1 inhibitor of the present invention inhibits intracellular Chk1 function
[0362] To determine that a Chk1 inhibitor of the invention inhibits Chk1 function in a cell, the inhibitor can be tested in a cell-based molecular assay. Since mammalian Chk1 has been shown to phosphorylate Cdc25C in vitro, suggesting that it negatively regulates cyclinB / cdc2 in response to DNA damage, Chk1 inhibitors were assayed for their ability to enhance cyclinB / cdc2 activity. The experiment can be designed as follows: BeLa cells are irradiated at 800 rads and incubated at 37°C for 7 hours. Because these cells are negative for p53 function, they only arrest at G2. Next, pyridazole was added to a concentration of 0.5 μg / mL, and the cells were incubated at 37° C. for 15 hours. The purpose of adding pyridazole is to capture any cells that cross the G2 arrest into M. Finally, Chk1 inhibitor was added for 8 hours as suggested by the manufacturer, cells were harvested, lysed and an equiva...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com