High-throughput single nucleotide polymorphism detecting method based on magnetic nano-particles and universal label technique
A magnetic nanoparticle, high-throughput technology, applied in the field of genetic testing, can solve the problems of high price and increase the cost of a large number of loci typing, and achieve the effect of reducing costs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment approach I
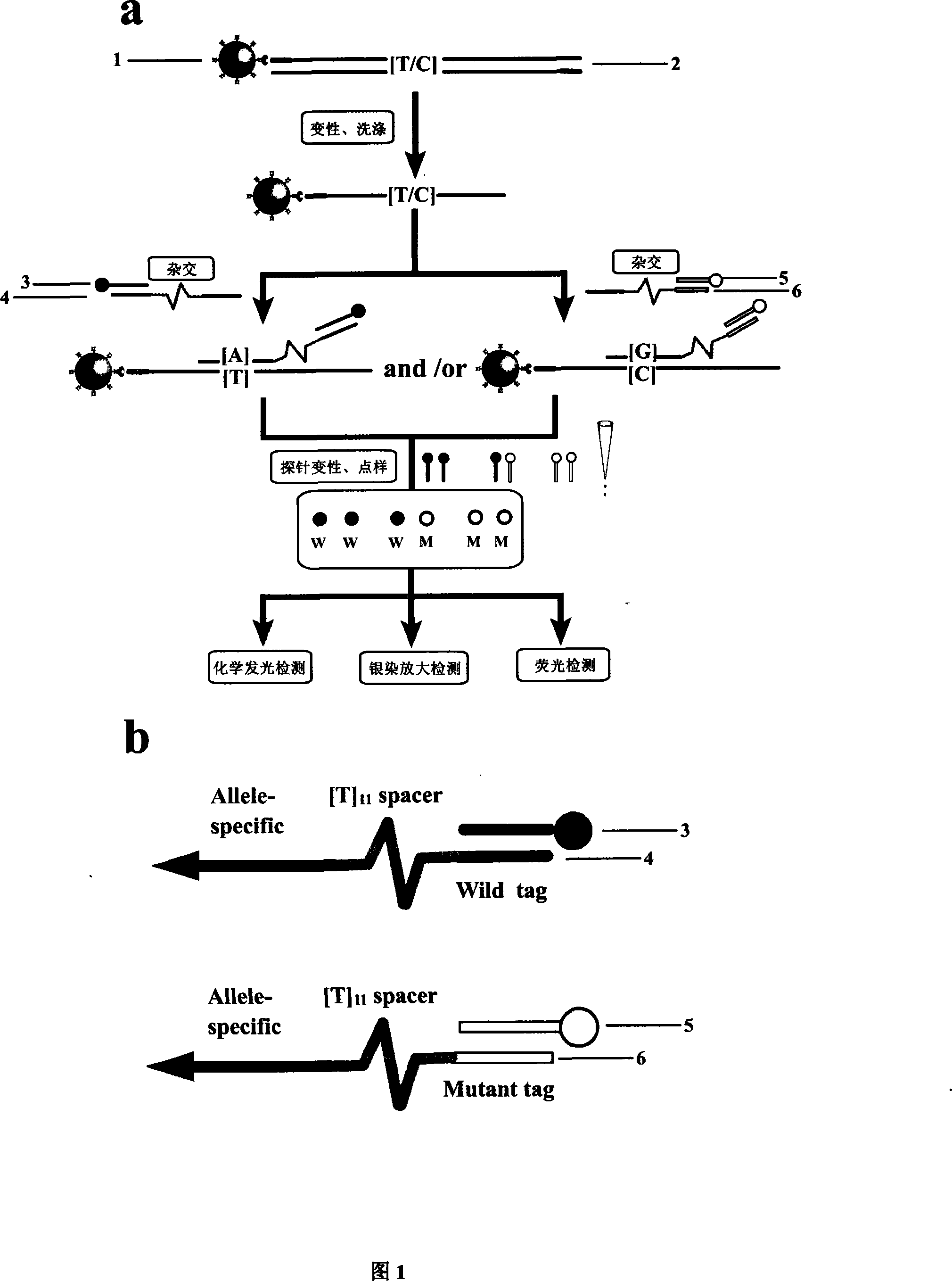
[0030]The invention relates to a method for high-throughput SNPs typing by using magnetic nanoparticles as carriers and universal label technology. The details are as follows: 1. Select several important functional SNP sites, design corresponding primers (one side of the primer is biotin-labeled), a pair of wild and mutant detection probes 4, 6 (sequences include those complementary to the target sequence, etc.) Gene-specific sequence, a Poly T spacer sequence with 11 bases in the middle and a universal label sequence) and a pair of labeled (fluorescent, nano-gold, luminescent enzyme) universal label detection probes 3, 5 (general The length of the detection probe is about 13 bases, which is complementary to the universal label sequence of the detection probe). 2. Amplify the biotin-labeled target sequence containing the SNPs site to be tested by PCR. The biotin-labeled target sequence can be the product of conventional PCR amplification, or it can be solid-phase amplification...
Embodiment approach II
[0032] The details are as follows: 1. Select several important functional SNP sites, design corresponding primers (one side of the primer is marked), a pair of wild and mutant detection probes 4, 6 (sequences include alleles complementary to the target sequence) specific sequence, a Poly T spacer sequence of 11 bases in the middle and a universal label sequence) and a pair of two-color fluorescent-labeled (wild-type Cy3, mutant Cy5) universal label detection probes 3 and 5 (universal detection probes The length is about 13 bases, and it is complementary to the universal label sequence of the detection probe). 2. Amplify the marked target sequence containing the SNPs site to be tested by PCR. The marked target sequence can be the product of conventional PCR amplification, or the product of solid-phase amplification PCR on the surface of the magnetic nanoparticles. 3. If the labeled target sequence 2 is a conventional amplification product, it is necessary to add an appropriate ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com