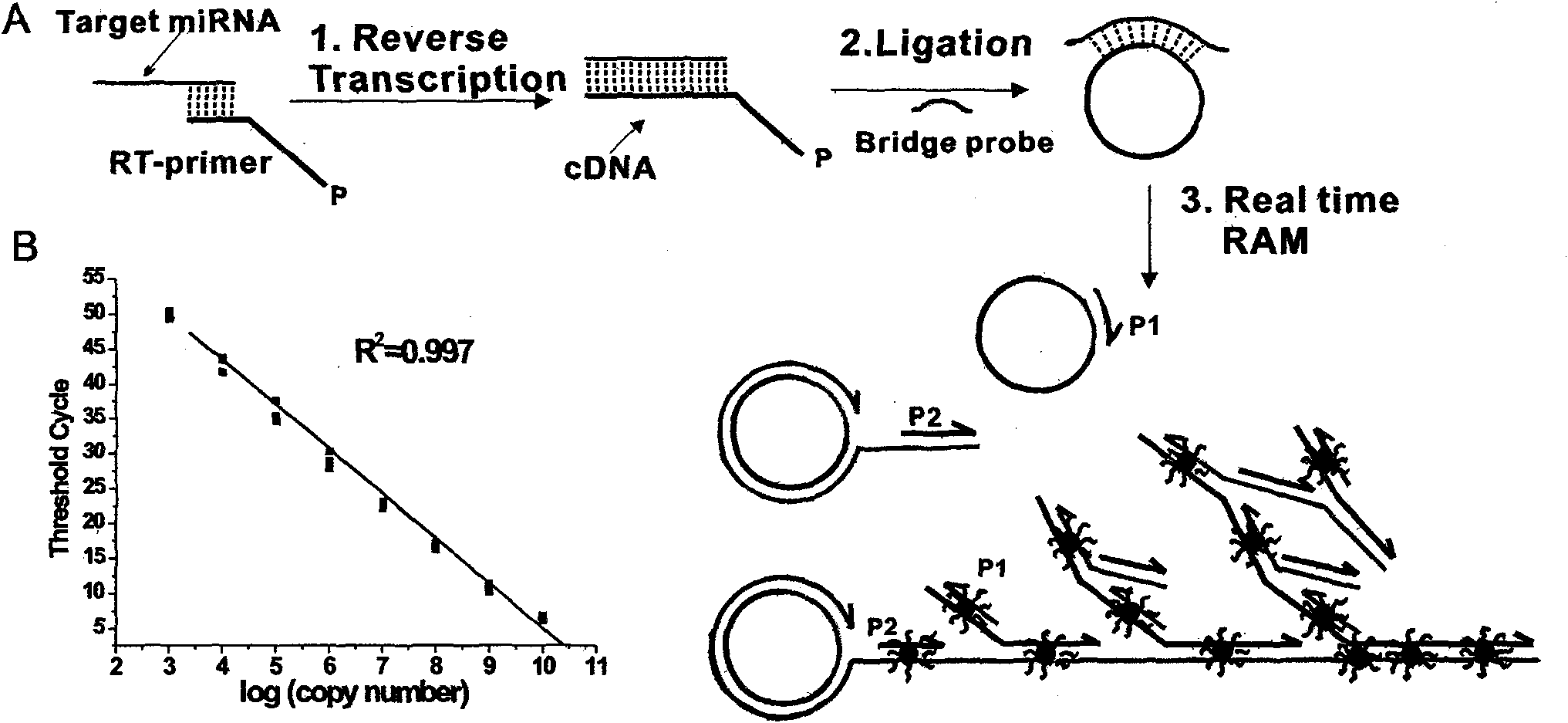
Method and kit for detecting small RNA
A kit and sequence technology, applied in the field of small RNA detection, can solve problems such as poor identification, and achieve the effects of strong specificity, high sensitivity and wide dynamic range
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0043] 3. Preparation of dsDNA template. 50 pmol of DNA oligo was annealed by incubation at 75° C. for 5 minutes, and then slowly cooled to room temperature (approximately 30 minutes). The reaction to prepare the dsDNA template was in the presence of 10 mM Tris HCl (Tris HCl), 50 mM NaCl, 10 mM MgCl 2 , 1 mM dithiothreitol (DTT), 25 μM dNTPs, and 5UKlenow Fragment (3'-5'exo) (New England BioLabs, Eastwin Scientific, China) in a 20-μL volume at 37°C for one hour. The reaction mixture was then heated to 75°C for 20 min to inactivate the enzyme and slowly cooled to room temperature to anneal the dsDNA.
[0044] 4. In vitro transcription reaction. 20 μL of the above dsDNA template mixture was added to 30 μL containing 0.5 mM NTPs, 40 mM Tris-HCl, 6 mM MgCl 2 , 10mM DTT, 2mM spermidine, 40-200U ribonuclease inhibitor (Ribonuclease Inhibitor (TAKARA, China, Dalian)) and 50 U T7 RNA polymerase (New England BioLabs, EastwinScientific, China) in the in vitro transcription buffer, T...
Embodiment 1
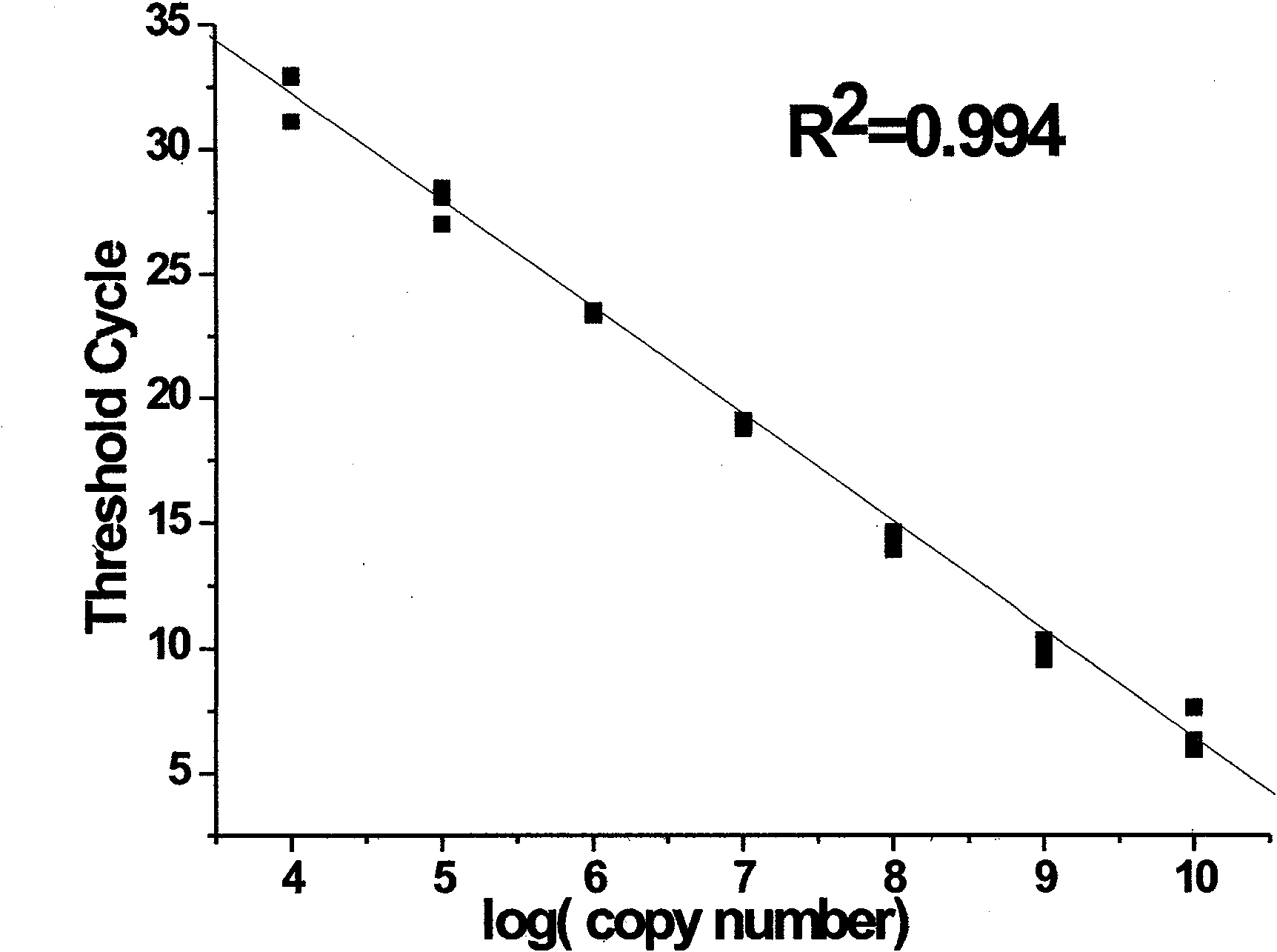
[0053] Detection of let-7a. The nucleotide sequence of let-7a is shown in Table 2, and the nucleotide sequences of RT-primers and bridge probes used in the detection process, as well as primer 1 and primer 2 in branch amplification, are shown in Table 3. The test results are shown in figure 1 b. Depend on figure 1 B It can be seen that the horizontal axis of the branch amplification curve (the common logarithmic value of the initial miRNA number) and the vertical axis C T The values showed a perfect linear relationship (R 2 = 0.997). As can be seen from the figure, the detection method of the present invention can detect as little as 10 3 (zeptomole) let-7a molecule and it has a dynamic range of at least 7 orders of magnitude for each reverse transcription reaction.
Embodiment 2
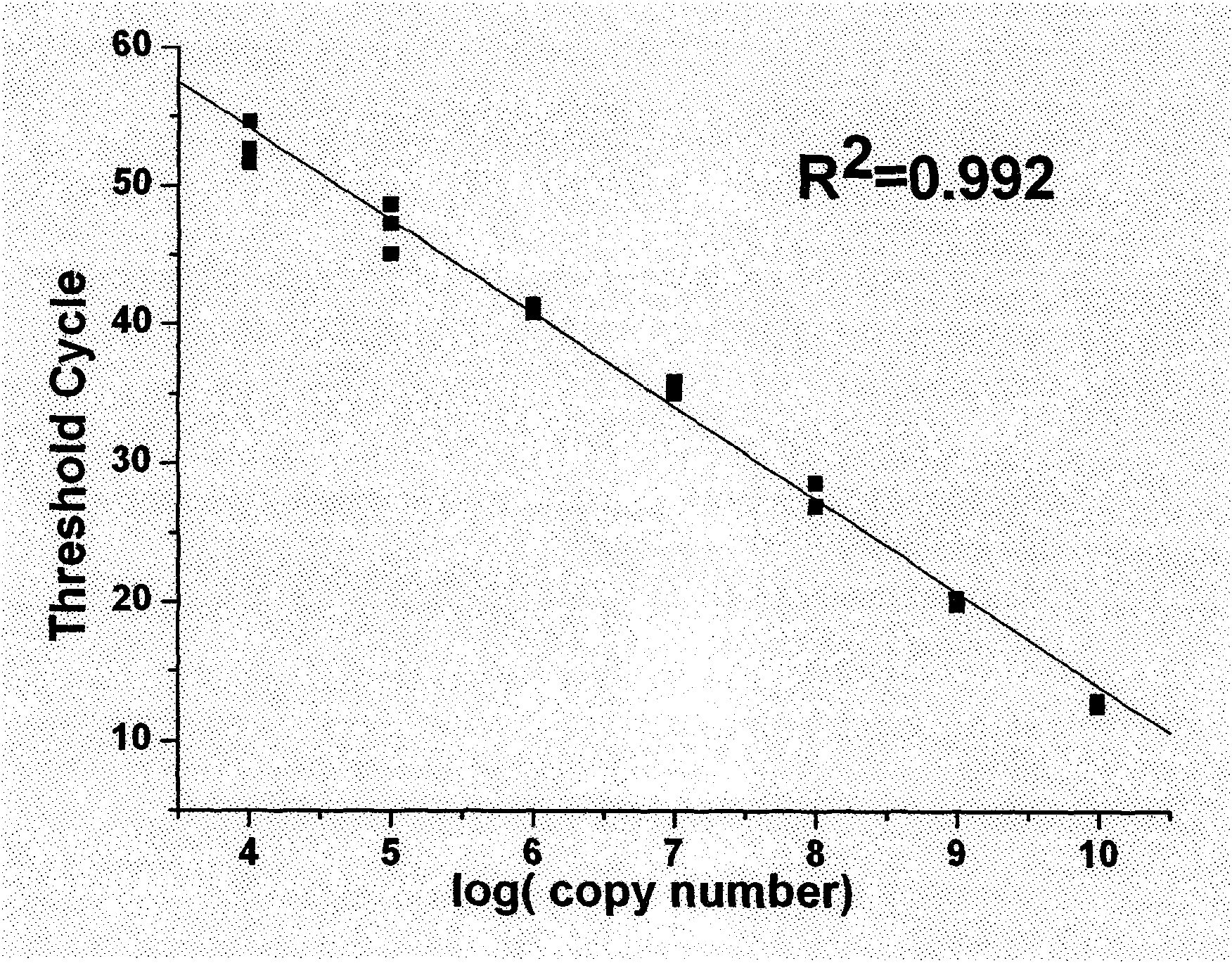
[0055] Detection of miR-1. The nucleotide sequence of mir-1 is shown in Table 2, and the nucleotide sequences of RT-primers, bridge probes used in the detection process, and primer 1 and primer 2 in branch amplification are shown in Table 3. The test results are shown in figure 2 . Depend on figure 2 It can be seen that the horizontal axis of the branch amplification curve (the common logarithmic value of the initial miRNA number) and the vertical axis C T There is a good linear relationship between the values (R 2 = 0.992). As can be seen from the figure, the detection method of the present invention can detect as little as 10 4 mir-1 molecules with a dynamic range of at least 6 orders of magnitude for each reverse transcription reaction.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com