Methods and compositions for breeding for preferred traits
A plant breeding and plant technology, applied in the direction of botany equipment and methods, biochemical equipment and methods, applications, etc., can solve the problems of high growth, large crop loss, expensive, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0242] Example 1: Genotyping of GLS responses
[0243] To detect QTLs associated with GLS resistance, plants were genotyped to determine GLS response. The rating scale below was used for phenotypic assessment of GLS and was used in all studies. The percentage of leaf area infected was used to rate plants on a scale from 1 (very high resistant) to 9 (susceptible). Disease resistance was assessed visually after pollination. Infection can be natural, or from artificial inoculation in experiments.
[0244] Table 1. Description of rating scales used for GLS phenotyping. ILA = infected leaf area.
[0245] illustrate
[0246] illustrate
Embodiment 2
[0247] Example 2: GLS Resistance Mapping Study 1
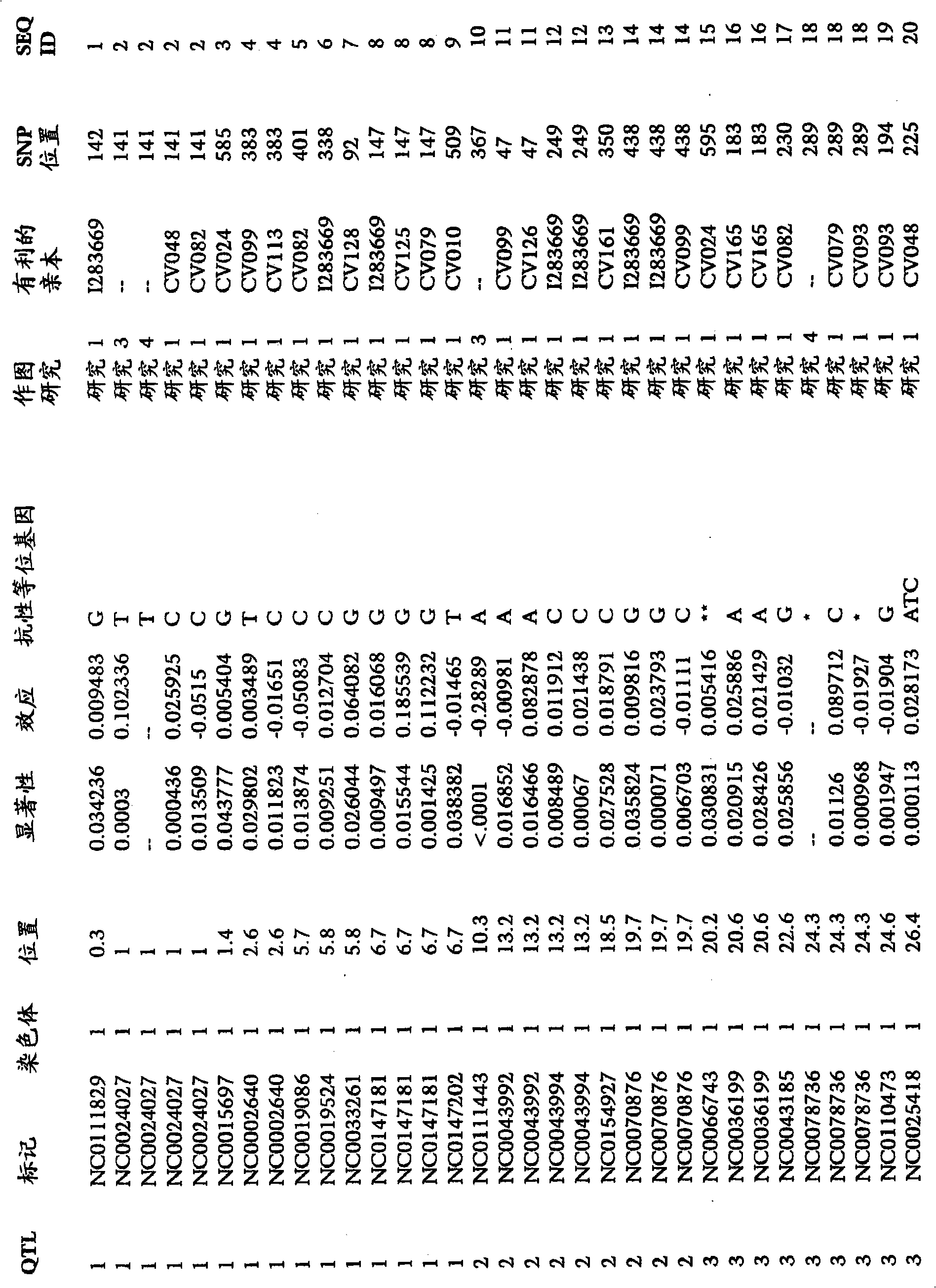
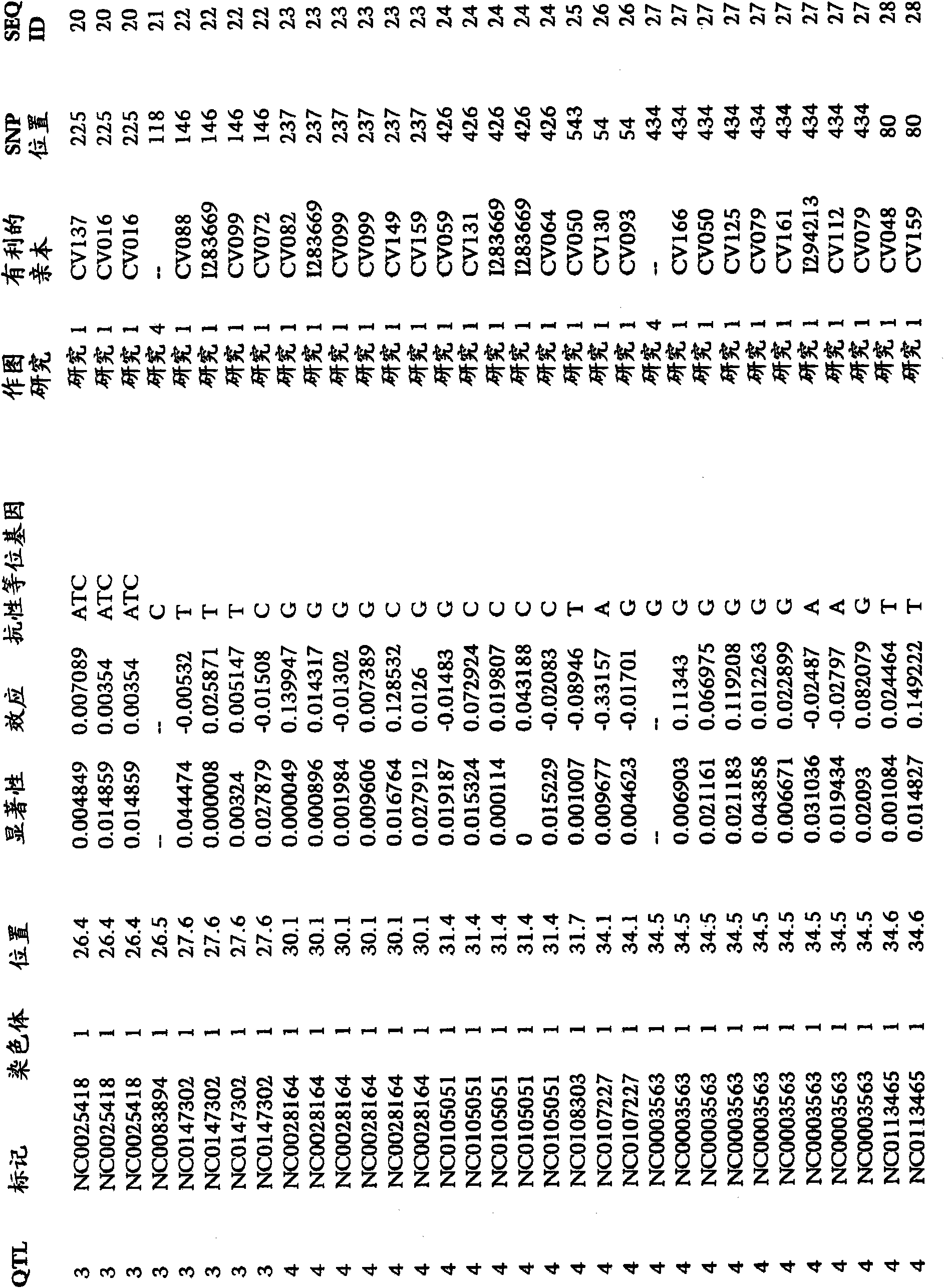
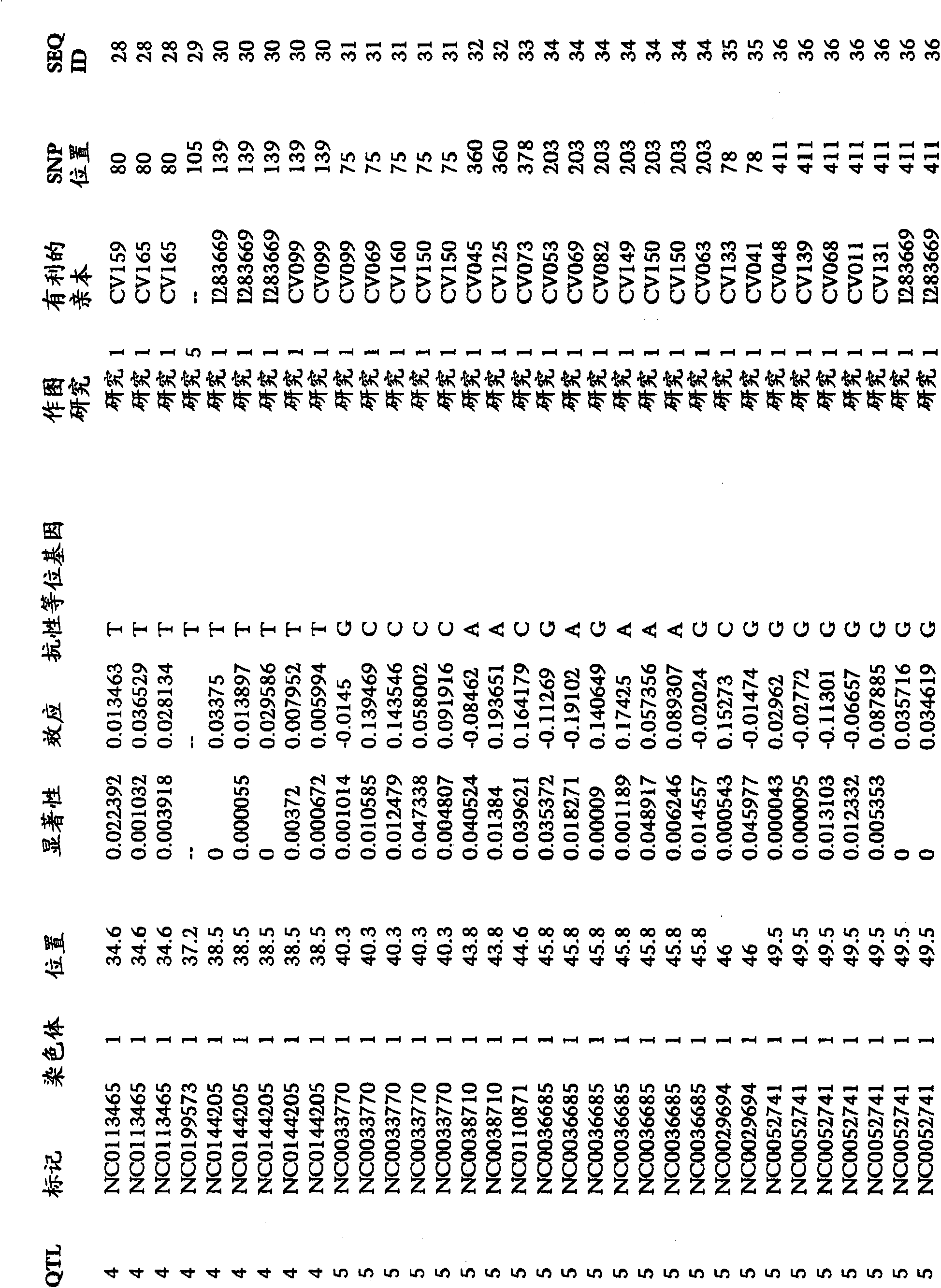
[0248] To examine the correlation between SNP markers and GLS resistance in maize, analytical data from many studies were combined. An association study was performed to assess whether there was a significant association between one or more marker genotypes and GLS resistance in one or more breeding crosses. The mapping study synthesized data from 176 mapping groups. The number of individuals in each population ranged from 95 to 276. The segregating populations were the following generations: F2, BC1F2, BC1 and DH. The number of SNP markers used for genotyping ranged from 55 to 158. Individuals were phenotyped for traits, including GLS resistance. A total of 2499 SNP markers on chromosomes 1, 2, 3, 4, 5, 6, 7, 8, 9 and 10 were identified for association with GLS resistance. The provided SNP markers can be used to monitor the introgression of GLS resistance into breeding populations. SNP markers, significance levels and f...
Embodiment 3
[0249] Example 3: GLS Resistance Mapping Study 2
[0250] An association study was performed to assess whether there was a significant association between one or more marker genotypes and GLS resistance in one or more breeding crosses. In the association study, 769 F2s from the CV128 / CV162 population were screened using 117 markers. Associations between a total of 53 SNP markers and GLS resistance were determined on chromosomes 1, 2, 3, 4, 5, 6 and 8. The provided SNP markers can be used to monitor the introgression of GLS resistance into breeding populations. SNP markers, significance levels and favorable alleles associated with GLS resistance in figure 1 report in.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com