Oligonucleotide retention time prediction method
An oligonucleotide and retention time technology, which can be used in biochemical equipment and methods, measuring devices, and microbial determination/inspection. big effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0013] The following is a detailed description of the use of the method of the present invention for oligonucleotide chromatographic retention time prediction as an example, including the following steps:
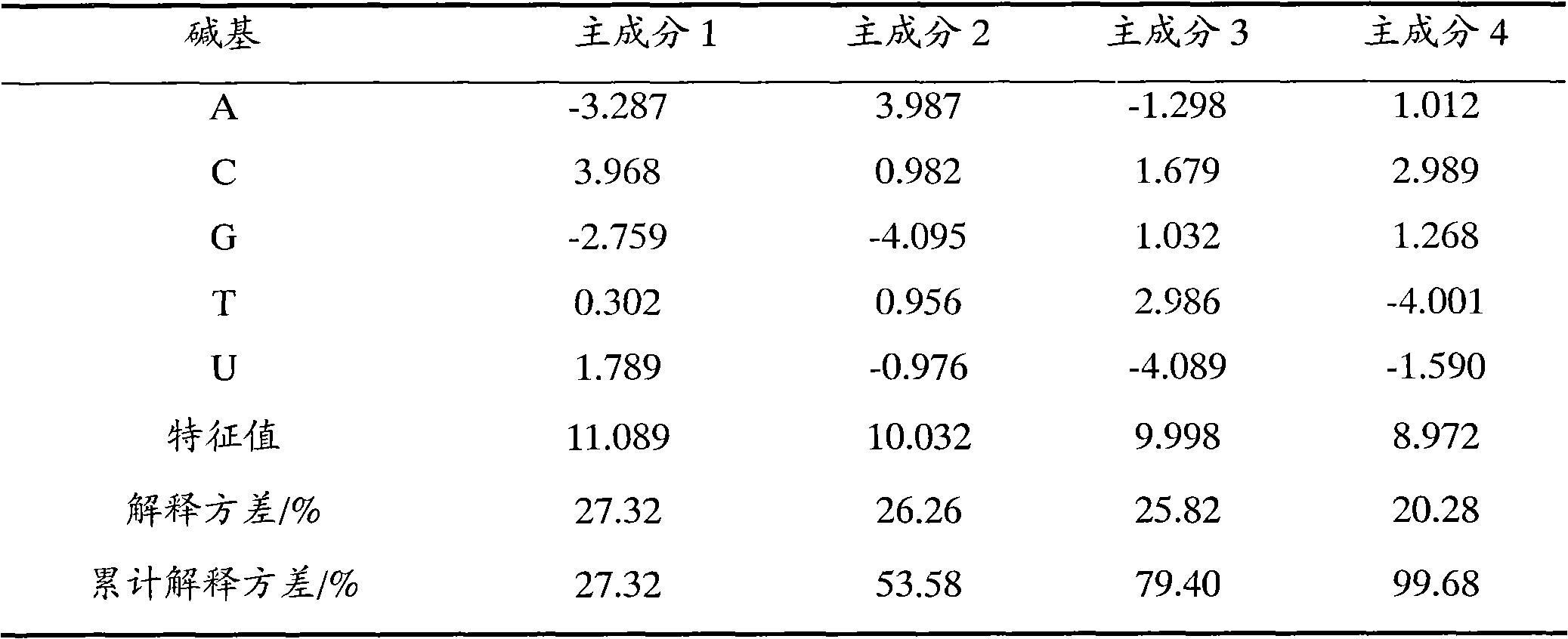
[0014] a) Select 585 three-dimensional property parameters of 5 kinds of bases (A, C, G, T and U), including: Randic molecular profile parameters, geometric feature parameters, radial basis function descriptors (RDF) based on different atomic distances ), the descriptor obtained by the molecular structure characterization (MoRSE) based on the electric diffraction method, the descriptor of the weighted whole invariant molecule (WHIM) and the set of geometry, topology and atomic weight (GETAWAY) parameters;
[0015] Principal component analysis is used to process 585 kinds of property parameters, and 4 principal components are obtained, which accumulatively explain 99.68% of the variance of the original data matrix (5×585). The scores of the principal components are shown in T...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com