Mycobacteria strain detecting dot blot membrane and kit
A detection kit and technology for mycobacteria, which is applied in the determination/inspection of microorganisms, microorganisms, biochemical equipment and methods, etc., and can solve problems such as easy missed detection, low pathogenicity, and small detection range
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
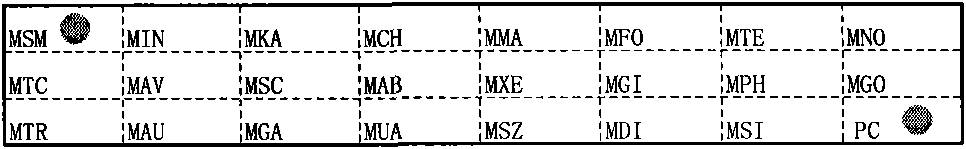
[0117] Embodiment 1 A kind of mycobacterium strain detection membrane strip
[0118] Comprising a substrate, and specific oligonucleotide probes immobilized on the substrate, the specific oligonucleotide probes include:
[0119] 1) A specific oligonucleotide probe for detecting Mycobacterium smegmatis, the base sequence of which is shown in SEQ ID NO: 1;
[0120] 2) A specific oligonucleotide probe for detecting Mycobacterium intracellulare, the base sequence of which is shown in SEQ ID NO: 2;
[0121] 3) a specific oligonucleotide probe for detecting Mycobacterium kansasii, the base sequence of which is shown in SEQ ID NO: 3;
[0122] 4) A specific oligonucleotide probe for detecting Mycobacterium chelonis, the base sequence of which is shown in SEQ ID NO: 4;
[0123] 5) A specific oligonucleotide probe for detecting Mycobacterium marinum, the base sequence of which is shown in SEQ ID NO: 5;
[0124] 6) A specific oligonucleotide probe for detecting Mycobacterium fortuitum...
Embodiment 2
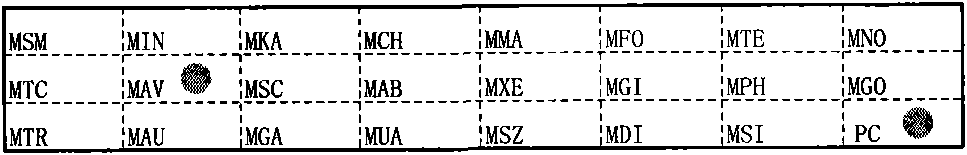
[0143] Embodiment 2 A kind of mycobacterium strain detection kit
[0144] The kit includes: a nylon membrane immobilized with a specific oligonucleotide probe and
[0145] A primer for specifically amplifying the insertion sequence between mycobacterial 16S rRNA and 23S rRNA, the base sequence of which is shown in SEQ ID NO: 24-25;
[0146] Specific oligonucleotide probes immobilized on nylon membrane strips include:
[0147] 1) A specific oligonucleotide probe for detecting Mycobacterium smegmatis, the base sequence of which is shown in SEQ ID NO: 1;
[0148] 2) A specific oligonucleotide probe for detecting Mycobacterium intracellulare, the base sequence of which is shown in SEQ ID NO: 2;
[0149] 3) a specific oligonucleotide probe for detecting Mycobacterium kansasii, the base sequence of which is shown in SEQ ID NO: 3;
[0150] 4) A specific oligonucleotide probe for detecting Mycobacterium chelonis, the base sequence of which is shown in SEQ ID NO: 4;
[0151] 5) A s...
Embodiment 3
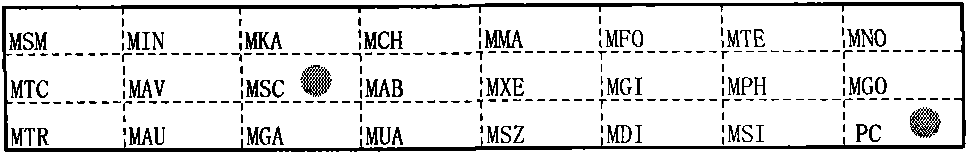
[0171] Clinical samples were detected with the detection kit described in Example 2,
[0172] Reagents required:
[0173] 20×SSC pH 7.0
[0174] NaCl 175.3g
[0175] Sodium citrate 88.2g
[0176] Add H 2 O 750mL was dissolved, and the pH was adjusted to 7.0 with a pH meter, and finally the volume was adjusted to 1000mL, and stored under high pressure sterilization.
[0177] 10% SDS pH 7.0
[0178] SDS 20g
[0179] Add H 2 O 180mL was dissolved, and the pH was adjusted to 7.0 with a pH meter,
[0180] Finally, the volume was adjusted to 200mL.
[0181] Liquid A (1×SSC, 0.1% SDS, pH 7.4)
[0182] 20×SSC 100mL
[0183] 10% SDS 10mL
[0184] Add H 2 O was adjusted to 1000mL.
[0185] Solution B (0.5×SSC, 0.1% SDS, pH 7.4)
[0186] 20×SSC 25mL
[0187] 10% SDS 10mL
[0188] Add H 2 O was adjusted to 1000mL.
[0189] Solution C (0.1M Sodium Citrate, pH 5.4)
[0190] 1M sodium citrate 100mL
[0191] Add H 2 O was adjusted to 1000mL.
[0192] Chromogenic solution...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com