Non-aggregating human VH domains
A domain, non-aggregation technology, applied in the field of non-aggregating human VH domains
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0139] Example 1: HVHP430V H library construction
[0140] Construction of fully synthetic phagemid-based human V HPhage display library.
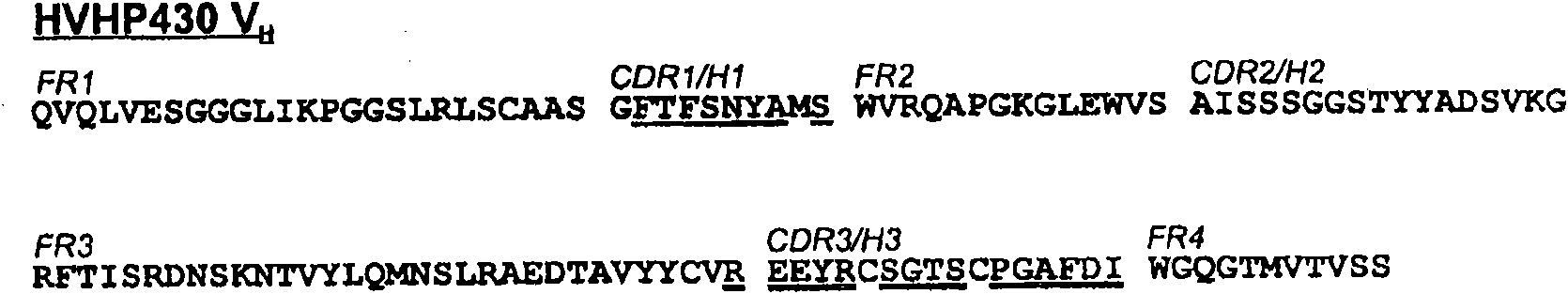
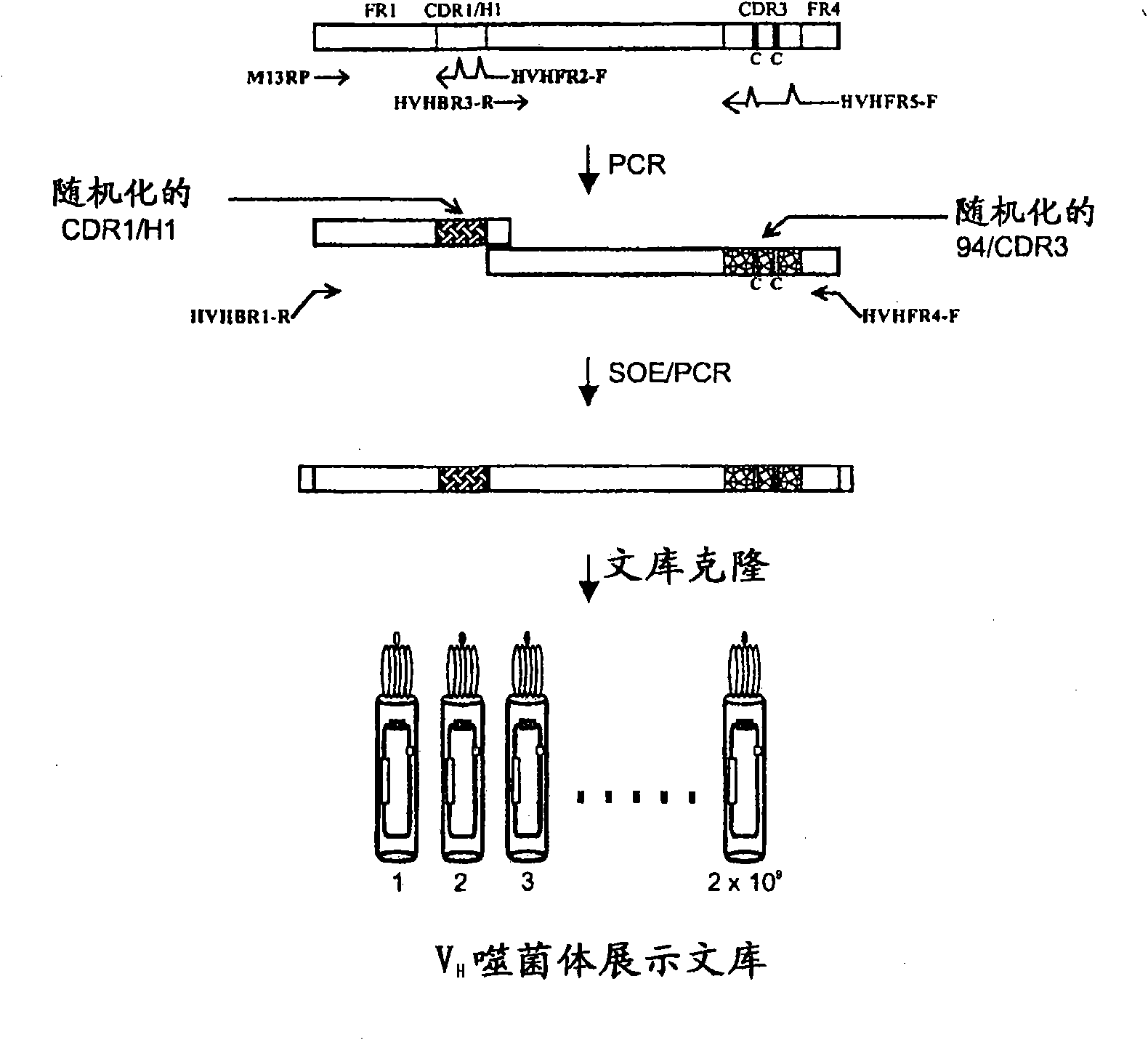
[0141] When in the HVHP430 bracket ( Figure 2A , SEQ ID NO: 1) build V H When using a library, maintain 2 CDR3 Cys to promote the formation of disulfide bonds within the CDR, thereby increasing the enzyme inhibitory V in the library H Frequency of. Randomize the remaining 14 CDR3 sites, site 94, and 8 H1 / CDR1 sites ( Figure 2A ). CDR2 was left unchanged as it has been shown to be involved in protein A binding (Randen et al., 1993; Bond et al., 2003). In addition, camelid VNARs that lack CDR2 (Stanfield et al., 2004) or utilize their CDR1 and CDR3 (Decanniere et al., 1999) or only CDR3 (Desmyter et al., 2001) for antigen recognition H H shows nanomolar affinities. according to Figure 2B The protocol shown in uses a phagemid vector ( image 3 ) to construct the library.
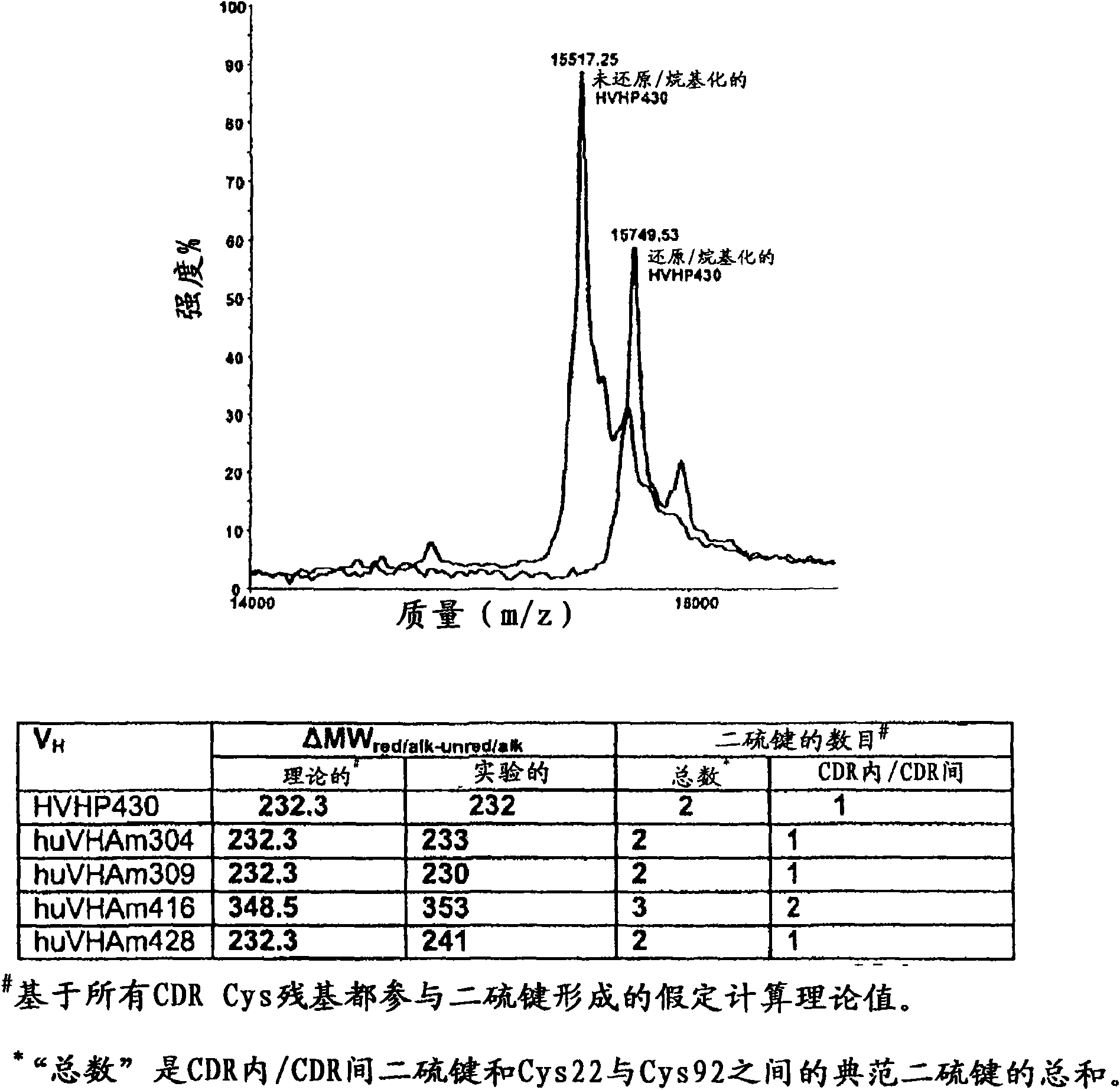
[0142] General V H HVHP430 (which has 2 Cys residues ...
Embodiment 2
[0148] Example 2: Panning and phage ELISA
[0149] A. Panning in the form of unit price display
[0150] In the first panning attempt, helper phage M13KO7 was used for superinfection, resulting in V H Monovalent display on the surface of phage (O'Connell et al., 2002). The initial aim was to explore the feasibility of the library in generating enzyme inhibitors. Four rounds of panning were performed against alpha amylase, lysozyme and carbonic anhydrase as described below.
[0151] A total of 50 μg of antigen (lysozyme, alpha amylase and carbonic anhydrase) in 100 μL PBS will be used to coat Maxisorp TM wells (Nunc, Roskilde, Denmark) overnight at 4°C. The solution was then removed and the wells were blocked by adding 200 μL of 3% BSA in PBS and incubating the wells at 37°C for 2 hours. Remove the blocking reagent and add 10 12 library phage (input), the wells were incubated at 37°C for 2 hours. The supernatant was removed and the wells were washed 7 times with 0.1% PBS...
Embodiment 3
[0168] Example 3: Analysis of clones
[0169] Nine V H , huVHAm302, huVHAm304, huVHAm309, huVHAm315, huVHAm316, huVHAm416, huVHAm427, huVHAm428 and huVHAm431 were used for subcloning and further analysis. However, except for 1 V H (huVHAm431), all V H Both have an amber stop codon that prevents their expression even in amber suppressor strains such as TG1 to be used as expression hosts (in TG1 cells the amber stop codon is read as an amino acid but mostly as a stop codon) . Therefore, the amber codon was replaced with a non-stop codon encoding the same amino acid before expressing the resulting V H .
[0170] However, inconsistent information was found regarding the nature of the amino acid encoded by the amber codon in E. coli TG1 cells when choosing an appropriate replacement codon. Some reported Glu as an overwriting amino acid (Hoogenboom, H.R. et al., 1991) (Baek, H. et al., 2002), while others reported Gln as an overwriting amino acid (Soltes, G. et al., 2003) (Ma...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com