Primers and method for detecting and typing human papilloma viruses in esophagi
A technology of HPV virus and esophagus, which is applied in the field of primers and methods for detecting and typing esophageal HPV virus, can solve problems such as application limitations, achieve the effect of reducing the use of consumables, simple and stable reagent consumables, and eliminating false positives
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
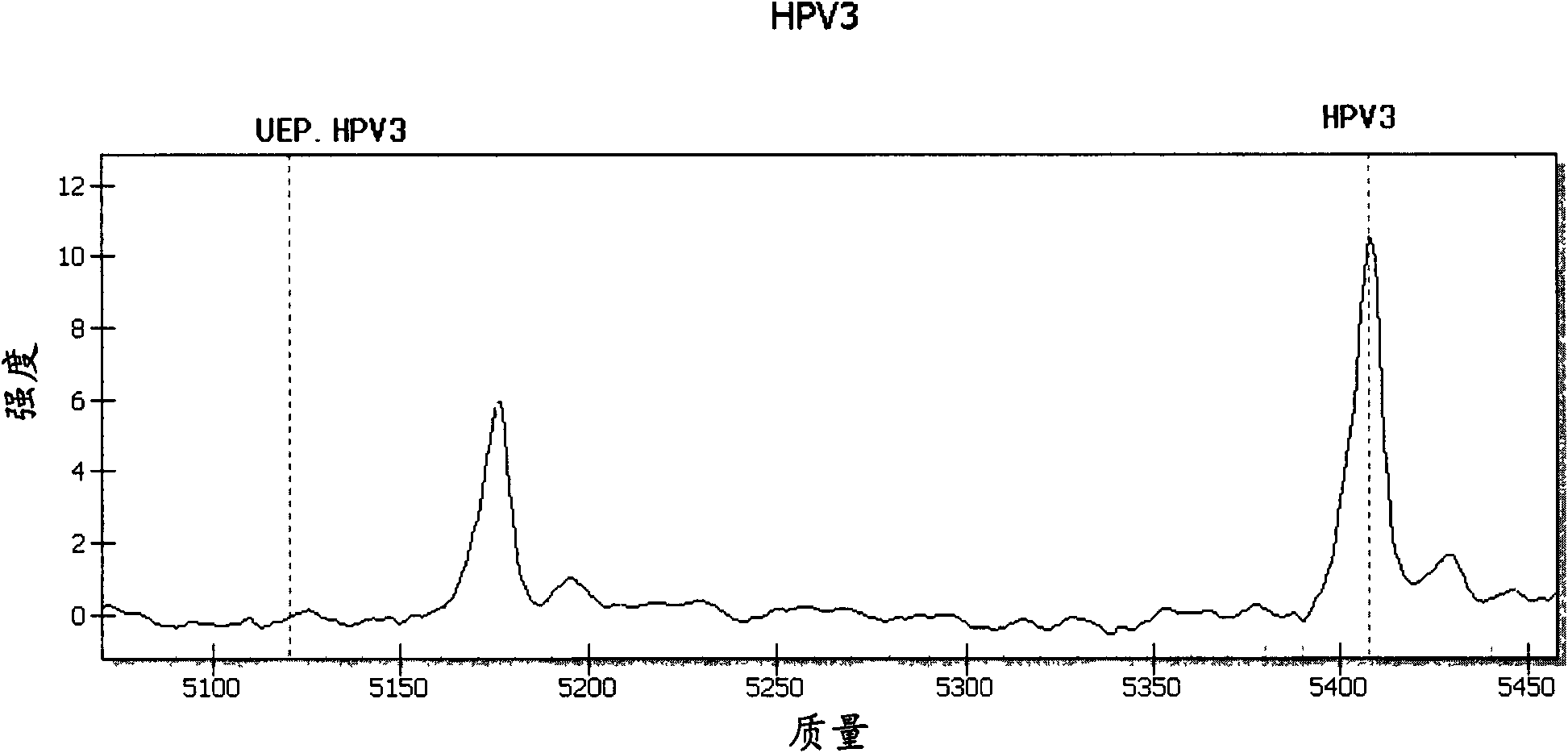
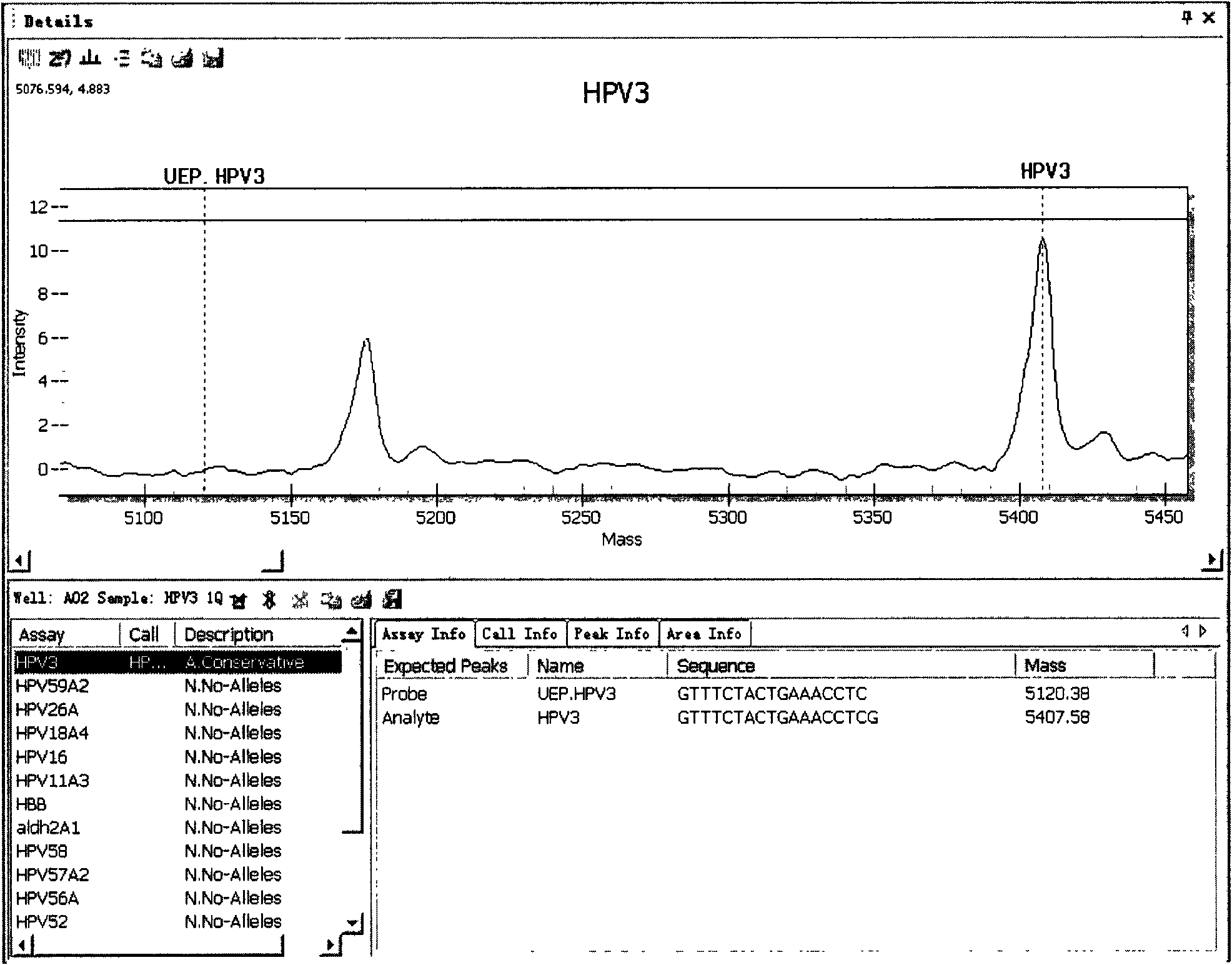
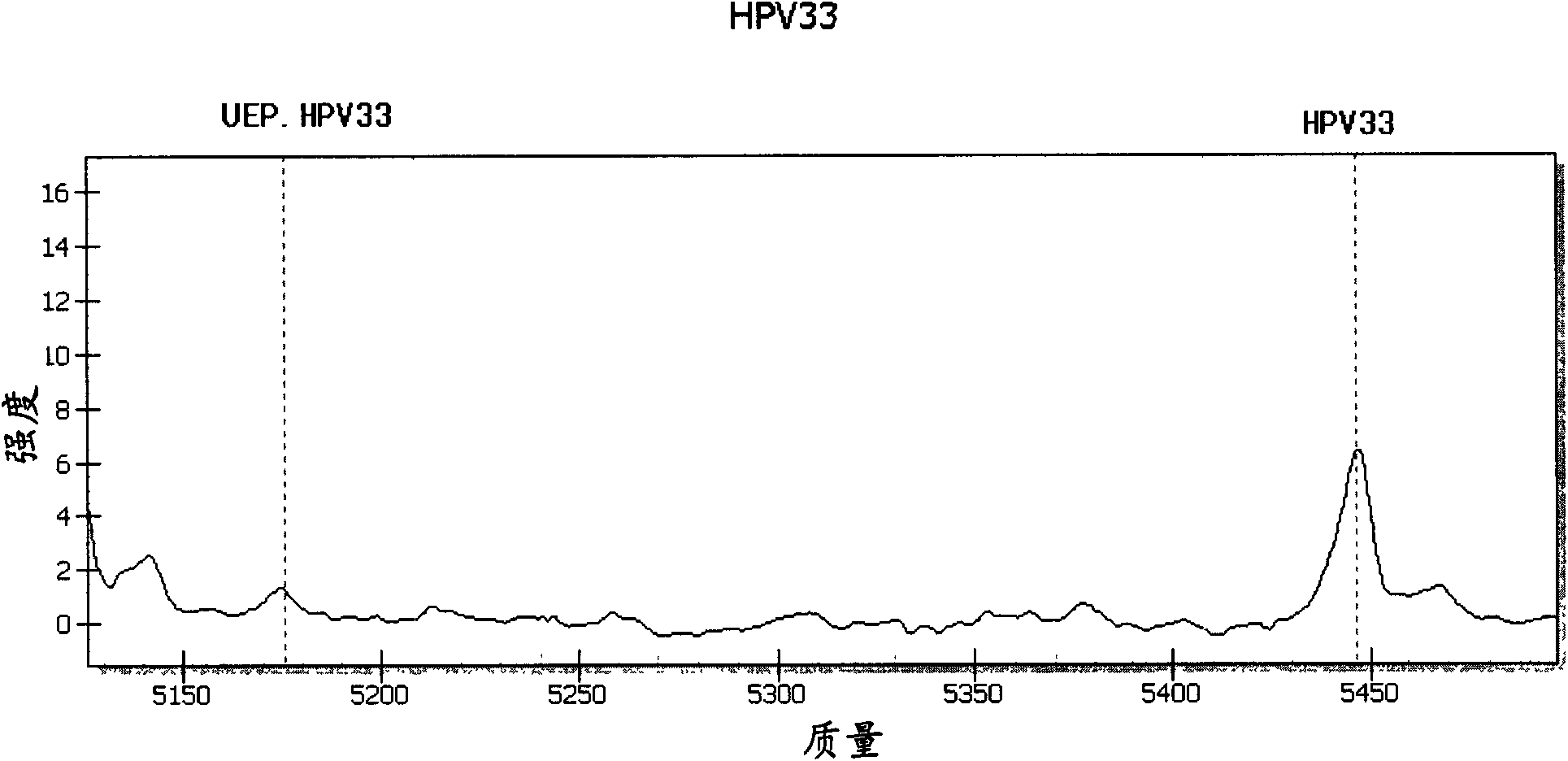
[0044] The embodiment of the present invention provides a method for detecting and typing the above 15 HPV types that infect esophageal cells, which includes the following steps.
[0045] (1) According to the gene sequences of the selected HPV types to be detected (that is, the above-mentioned 15 HPV types), on the basis of GP5+ / GP6+ universal primers, design amplification primers specific to each type, so The amplification primer has 12 bases that completely match the gene sequence of the type it is targeting at the 3' end, and has a tag sequence of 10 bases (acgttggatg) at the 5' end; the extension primer is designed, and the extension primer The length is 17-28 bases, and contains a type-specific polymorphic site marker in the 4 bases at its 3' end including the extended base, and the position selected for the extended primer is GP5+ / A relatively conserved region in the GP6+ sequence; wherein, the molecular weight difference between the extension product and the extension ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com