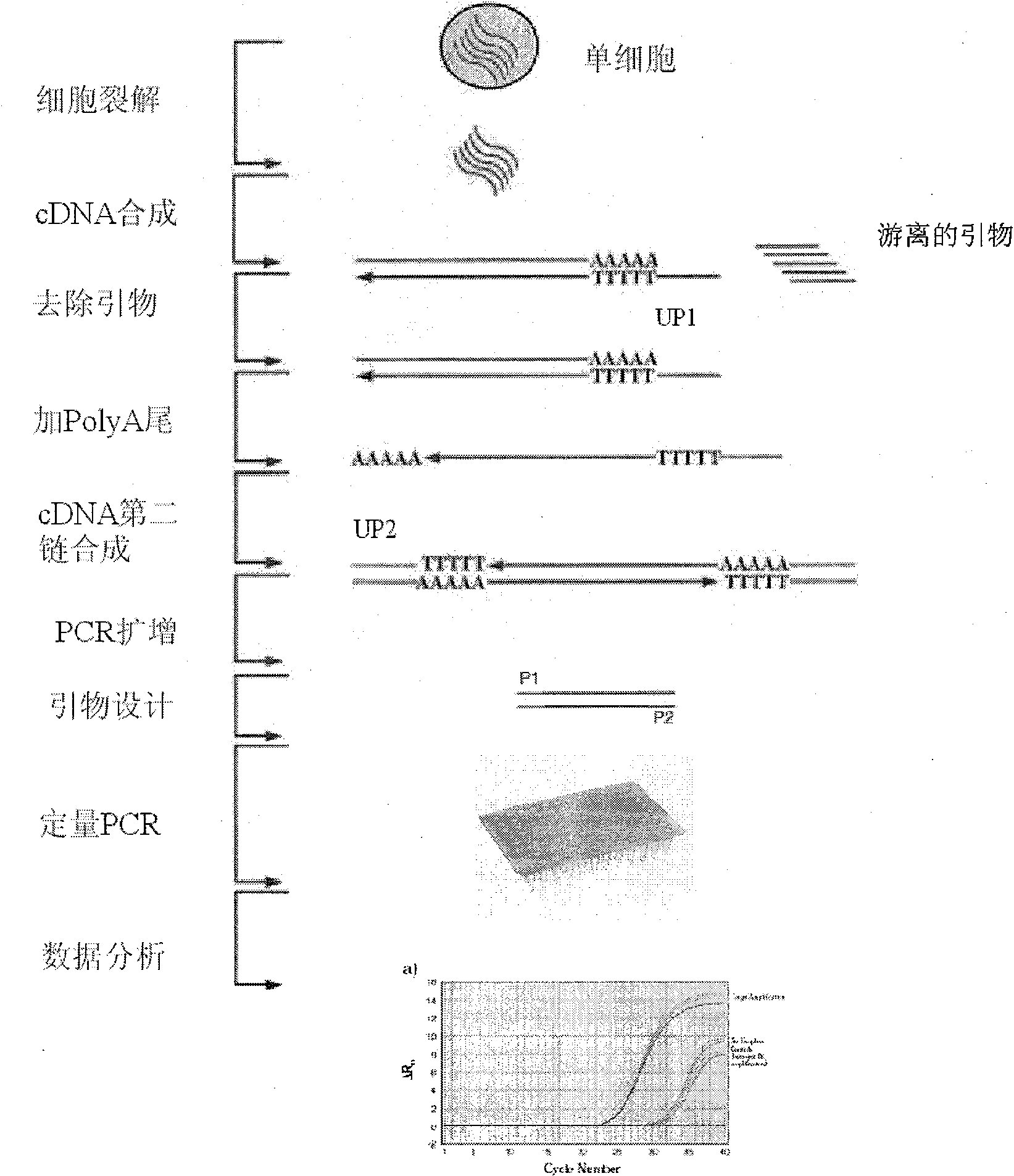Preimplantation genetic diagnosis on embryo by using new single cell nucleic acid amplification technology
A single-cell and embryonic technology, applied in biochemical equipment and methods, microbial measurement/inspection, etc., can solve the problems of expensive probes and lack of intellectual property rights, and achieve the effect of cheap price and convenient use
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment approach
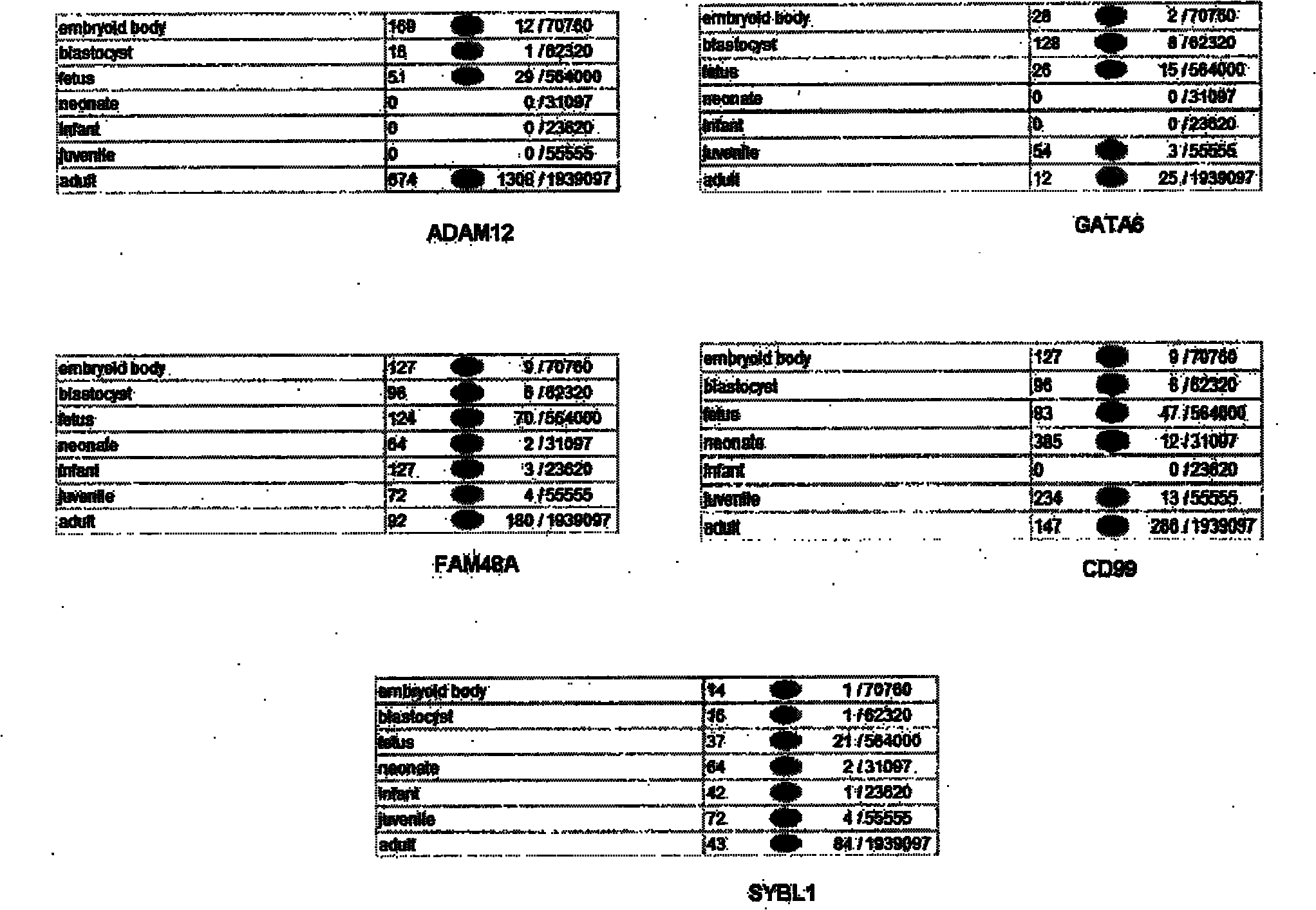
[0055] 3. Specific gene selection: the method for specific gene selection is detailed in the summary of the invention, and the nucleic acid sequence is obtained from the nucleic acid professional database according to the gene shown in Table 1. The sequence contains the expression tag sequence (STS), and is designed with the primers of ABI company Software designed primers. For the specific nucleic acid sequence of each gene used, please refer to the later part of the specification.
[0056] 4. Using SYBR as a fluorescent dye, of course, quantitative PCR with TaqMan probes can also be used. Here, SYBR is mainly cheap, convenient, and the accuracy meets the requirements. Use a 96-well plate for quantitative PCR (30-35 cycles), use GAPDH and OCT4 as reference genes; use normal male and female single embryos as controls, compare the quantitative PCR results of single-cell embryos to be tested with the control, Judging whether the chromosome to be detected is normal or not, so as...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com