Specific primers and kit for detecting various serotype pathogenic bacteria of legionella pneumophilia
A specific technology for Legionella pneumophila, applied in the field of multiplex PCR rapid detection, can solve the problems of unstandardized methods, poor stability of monoclonal antibody typing, and easy to be affected by environmental factors, etc., achieving simple method and promising market application wide, saving manpower and material resources
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] Example 1 : Extraction of the genome
[0042] 1) Take a little bacterial solution from the bacterial storage tube, streak it and inoculate it on the Legionella BCYE growth plate; 2.5% CO 2 , 37 ℃ cultured for 5-7 days.
[0043] 2) Add 1 mL of 50 mM Tris-HCl (pH 8.0) to the plate, scrape the bacteria with a sterilized coating rod, take an appropriate amount of bacterial liquid into a 1.5 mL centrifuge tube, centrifuge at 10,000 rpm for 5 minutes to accumulate bacteria, and remove the supernatant.
[0044] 3) Add 250 μL of 50 mM Tris-HCl (pH8.0) to resuspend, add 10 μL of 0.5M EDTA (pH8.0), and mix well.
[0045] 4) Add 15 μL of 20 mg / mL lysozyme, mix well, and incubate at 37°C for 20 minutes.
[0046] 5) Add 3 μL of 20 mg / mL proteinase K and mix gently.
[0047] 6) Add 20 μL of 10% SDS, water bath at 50°C for 1 hour until the solution is clear.
[0048] 7) Add 2 μL of 25mg / mL RNAase, and water bath at 65°C for 20 minutes.
[0049] 8) Add an equal volume of phenol:...
Embodiment 2
[0054] Example 2: Design of primers
[0055] The sequences downloaded from NCBI, combined with laboratory self-test sequences, were designed for the specific regions of the wzt genes of Legionella pneumophila types 1 and 6, and the wzm genes of Legionella pneumophila types 4, 10 and 13. The primer sequences are as follows: 1 shows:
[0056] Table 1 Specific primer sequences for Legionella pneumophila serotypes 1, 4, 6, 10 and 13
[0057]
Embodiment 3
[0058] Example 3 : Screening of specific primers
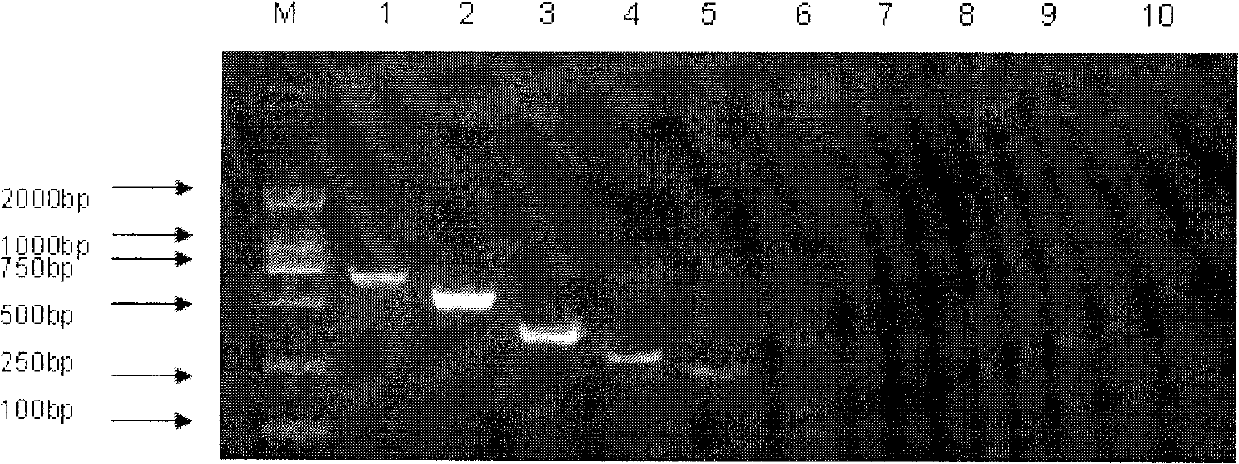
[0059] Primers were designed for the specific regions of the wzt genes of Legionella pneumophila types 1 and 6 and the wzm genes of Legionella pneumophila types 4, 10 and 13. The primer sequences are shown in Table 1 (SEQ ID NO: 1-SEQ ID NO: 10). Collected 1 standard strain of Legionella pneumophila type 1, 14 standard strains of other serotypes of Legionella pneumophila, 7 other strains of Legionella, 1 clinical isolate of Legionella pneumophila type 1, 4 Legionella The specificity of the primers was verified for clinical isolates of other serotypes and 6 closely related bacteria. The strain numbers and sources are shown in Table 2 below.
[0060] Table 2 Standard strains for testing
[0061]
[0062]
[0063]
[0064] The clinical strains used in this patent are shown in Table 3 below:
[0065] Table 3 Biochemical test results of tested clinical strains
[0066]
[0067] The PCR system is 0.3μl of 10uM prim...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com