Template-independent ligation of single-stranded DNA
A technology of RNA ligase and template, applied in the field of chemical improvement, can solve problems such as differences in intramolecular connection efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
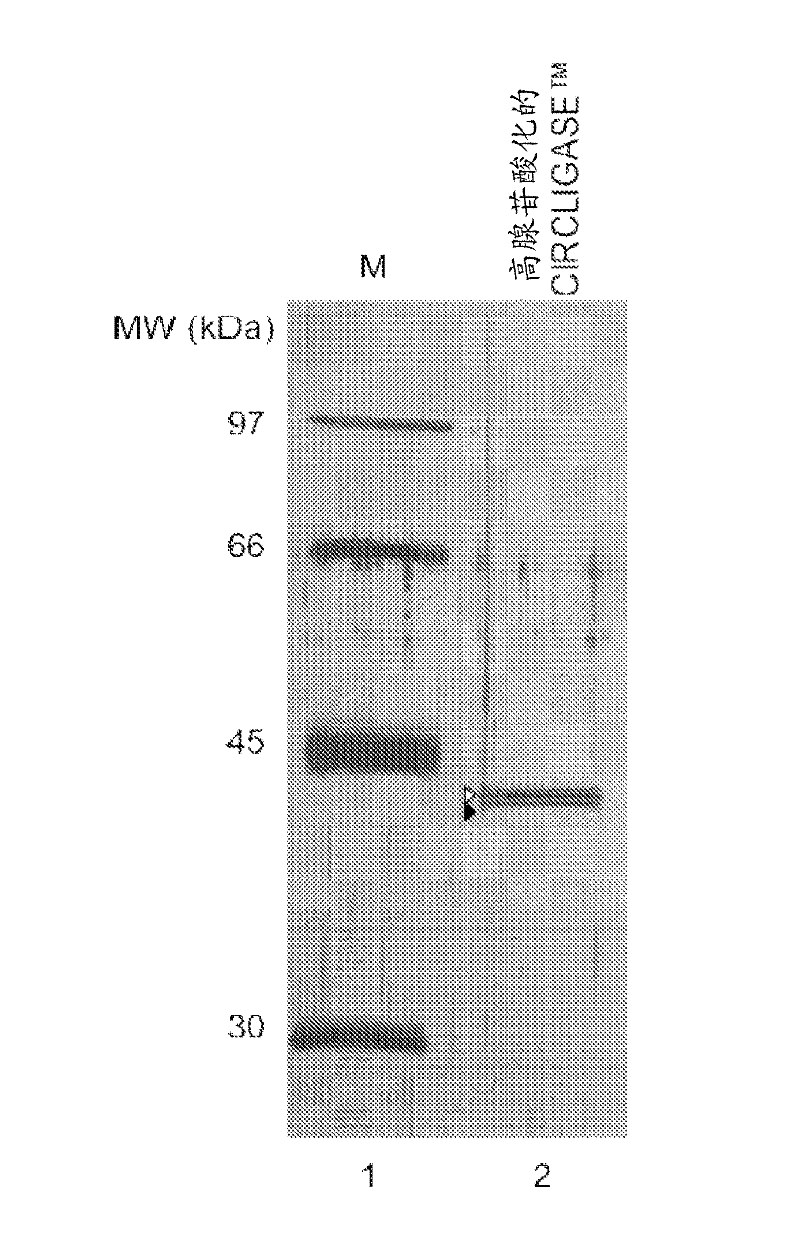
[0151] Purified CIRCLIGASE from a new clone of a thermostable RNA ligase gene from bacteriophage TS2126 TM Enzyme SDS-PAGE analysis
[0152] Adenylated CIRCLIGASE TM The enzyme migrated on a 10% SDS-PAGE gel as non-adenylated CIRCLIGASE TM Slightly taller strips. figure 1 Show the CIRCLIGASE obtained from the new clone TM Silver-stained SDS-PAGE gels of enzyme preparations. Percent adenylation was estimated by comparing the relative intensities of adenylated and non-adenylated bands; CIRCLIGASE from a new clone shown here TM The enzyme is estimated to consist of approximately 70% adenylated form and approximately 30% non-adenylated form.
Embodiment 2
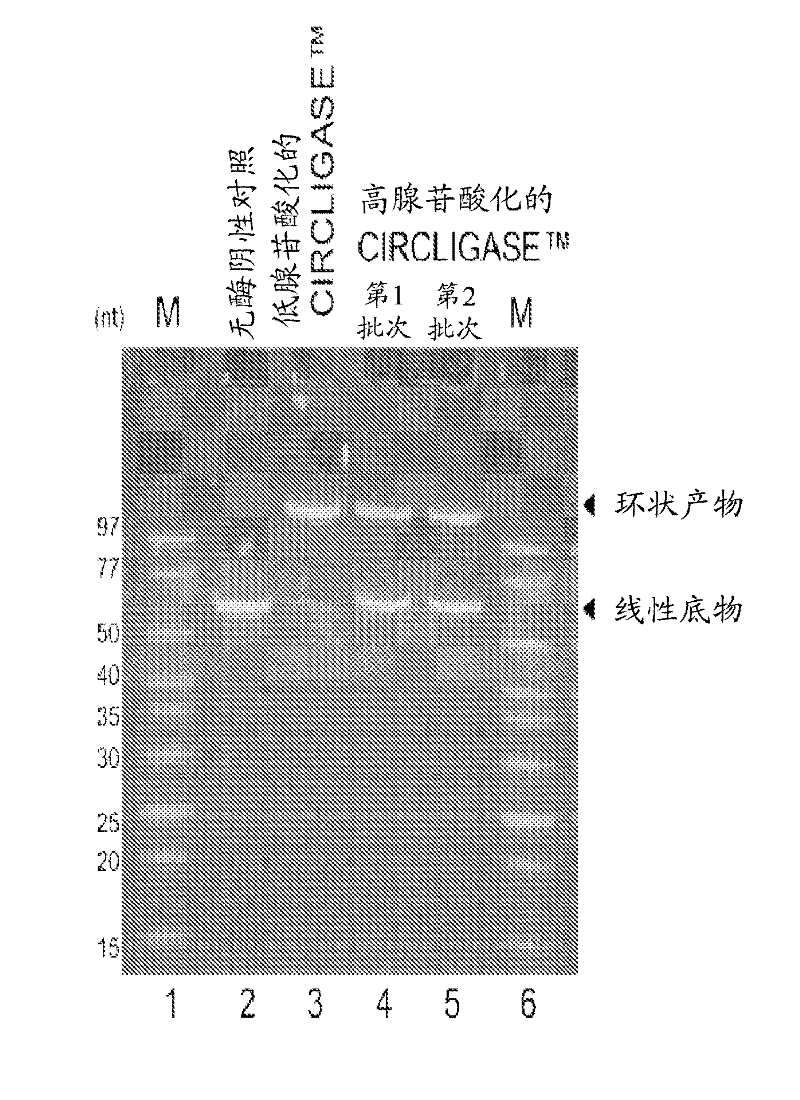
[0154] Hyperadenylated CIRCLIGASE from a new clone TM Enzymes with hypoadenylated CIRCLIGASE from old clones TM Comparison of Intramolecular Ligating Activities of Enzymes
[0155] Hypoadenylated CIRCLIGASE from old clones when assayed under standard ligation reaction conditions TM The enzyme converts approximately >95% of the 55-nucleotide control oligonucleotide (linear ssDNA supplied with CIRCLIGASE ssDNA Ligase) to circular ssDNA product ( figure 2 ). In contrast, the hyperadenylated CIRCLIGASE enzyme from the new clone converted only approximately 50% of the same linear ssDNA oligonucleotides to circular ssDNA products.
Embodiment 3
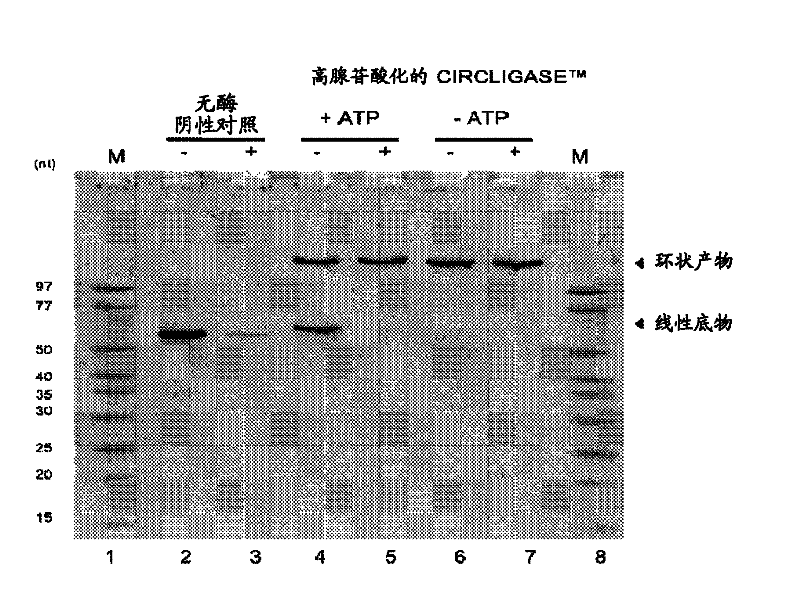
[0157] Effect of ATP concentration on the hyperadenylated form of CIRCLIGASE purified from a new clone TM Effect of ssDNA ligase on intramolecular ligation of linear ssDNA
[0158] In the presence of 50 μM ATP in the standard ligation reaction compound, approximately 50% of the 55-nucleotide ssDNA control oligonucleotide supplied with CIRCLIGASE ssDNA Ligase was converted to circular ssDNA product. When ATP was omitted from the standard ligation reaction mixture, approximately >95% of the 55-nucleotide ssDNA control oligonucleotide was converted to a circular ssDNA product ( image 3 ). The circular ssDNA product was resistant to digestion by 20 units of exonuclease I (Exo I; EPICENTRE), a single-strand specific exonuclease that requires a free 3' end. Unligated linear ssDNA substrates are degraded by Exo I, whereas circular ssDNA ligation products are resistant to Exo I digestion.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com