New method for positioning and cloning rice genes
A gene positioning and new method technology, applied in the field of biological gene positioning cloning, can solve the problems of low polymorphism in the target gene region, difficult to identify or clone genes, difficult to accurately locate or clone genes, etc., to reduce experimental consumption, The effect of improving the drawing efficiency and shortening the time course
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
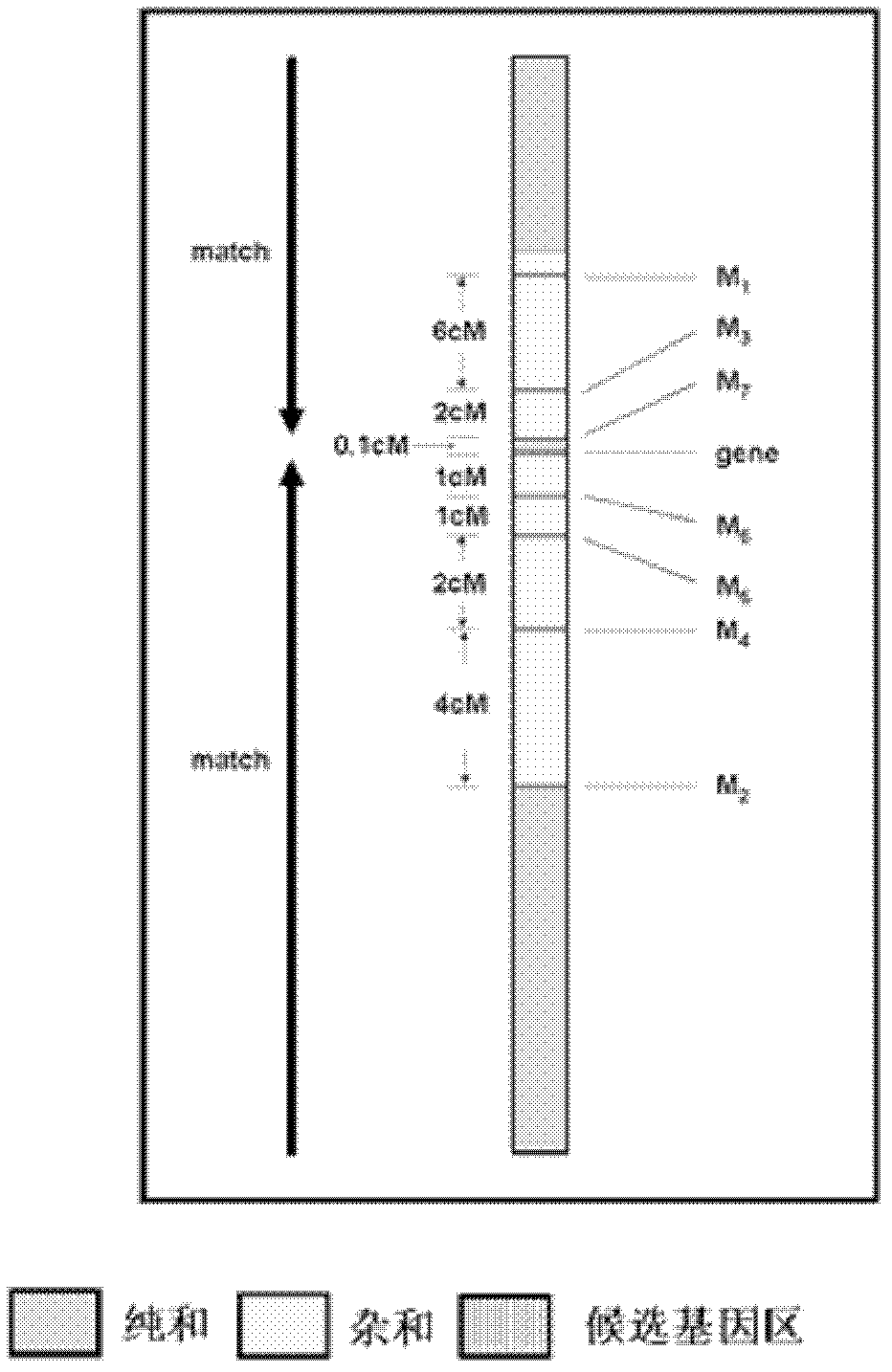
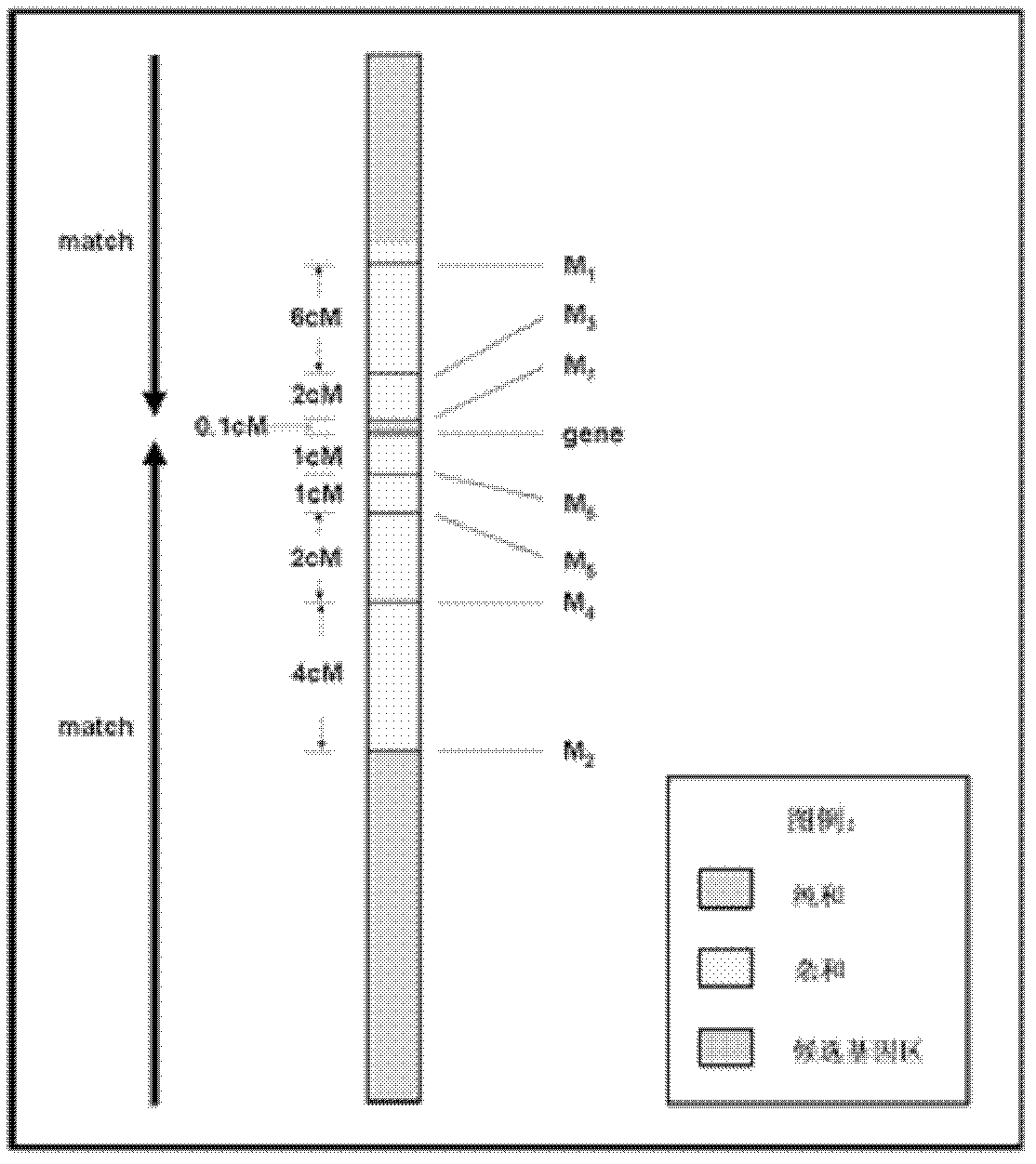
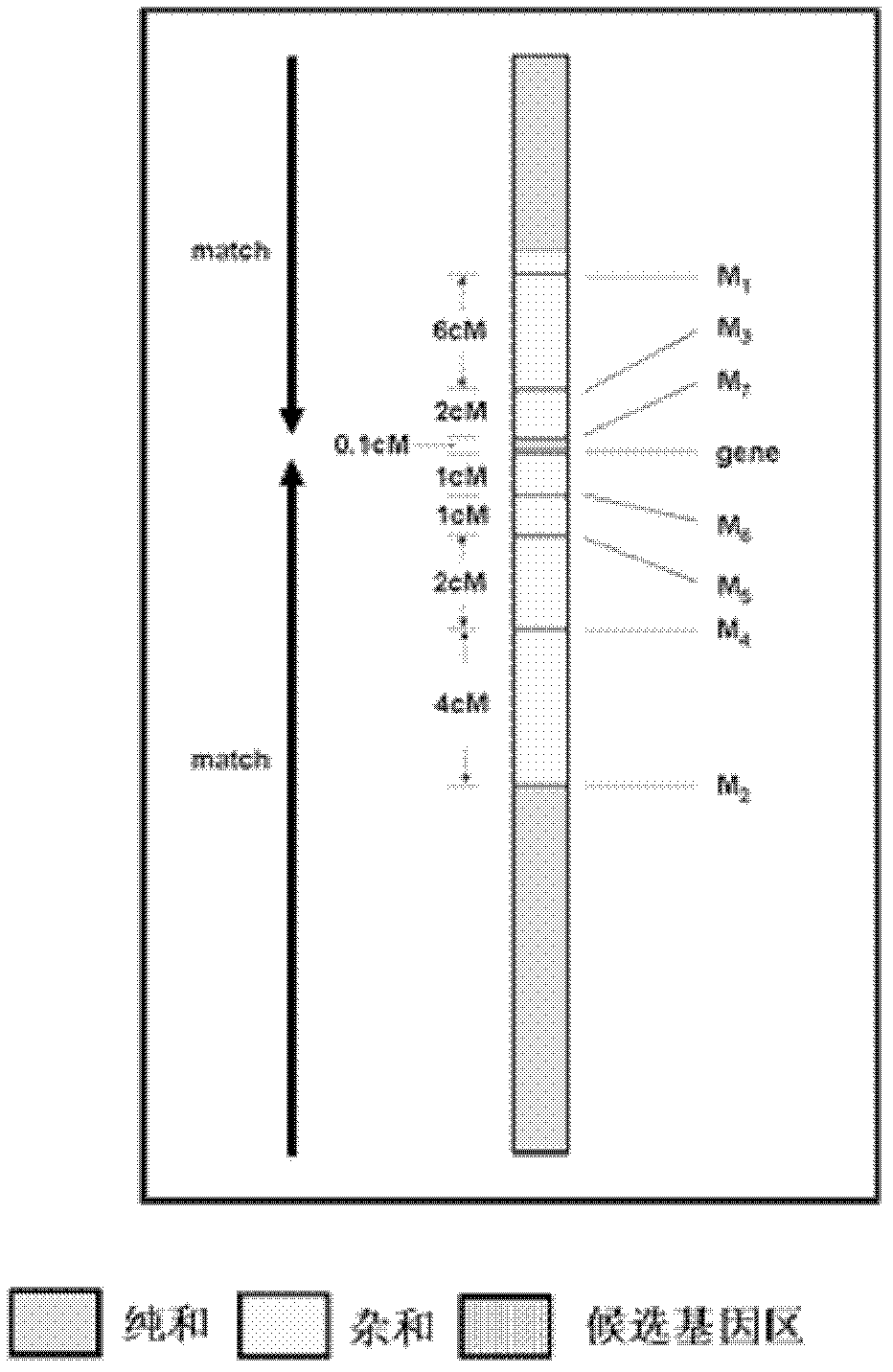
[0023] A new method for positional cloning of rice genes comprises the following main methods and steps:
[0024] (1) Construct a gene mapping group,
[0025] Two parents with high polymorphisms are used to cross or backcross to establish a gene mapping population. The population indicators are: 1) 80% polymorphism; 2) normal fertility; 3) less than 10% deviation from segregation sites.
[0026] (2) Using the F5 or BC2 F1 generation as the basic material, construct gene mapping populations, mapping populations (partial mapping), and gene effect populations.
[0027] (3) Guided by traits, the target gene region was determined by interval mapping as the basic method.
[0028] (4) Identify the target gene based on the "Match" principle.
[0029] (5) Establish multiple populations for identification of gene effects, with 2-3 populations (establish secondary populations if necessary).
[0030] (6) When the genetic distance between closely linked markers and genes is 0.1-1CM, sel...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com