Method for real-time quantitative polymerase chain reaction (PCR) detection of bifidobacteria and Escherichia coli by using Taqman probes
A bifidobacterium and Escherichia coli technology, applied in the field of molecular biology detection, can solve the problems of affecting quantitative results, poor accuracy, poor specificity, etc., and achieve the effect of solving PCR pollution, reducing non-specific amplification, and saving detection time
Inactive Publication Date: 2012-07-11
SHENZHEN CHILDRENS HOSPITAL
View PDF1 Cites 10 Cited by
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
[0003] When detecting bacteria, the most traditional method is to use traditional culture counting, but because there are a large number of unculturable microorganisms in natural environments, especially extreme environments, the unculturability of a large number of microorganisms is a reminder of the structure of microbial communities in nature by traditional microbial ecology. , ecological functions and their interrelationships are the biggest obstacles in the research
Bacterial pure culture detection method takes a long time, poor specificity, low sensitivity, poor accuracy and other shortcomings
[0004] The use of molecular biology methods to detect bacteria or viruses is currently a relatively common detection method, but ordinary PCR methods can only achieve qualitative or relative quantification, and cannot accurately quantify the target
SYBR GREEN fluorescent quantitative PCR method detects that SYBR Green binds to double-stranded DNA during the PCR reaction process. With the increase of PCR products, the amount of PCR products combined with SYBR Green also increases. Although the purpose of quantification can be achieved, intercalating dyes cannot Misidentified amplification products or primer-dimers generated during the reaction can affect quantitative results
And prone to false positives due to contamination during operation
Method used
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
View moreImage
Smart Image Click on the blue labels to locate them in the text.
Smart ImageViewing Examples
Examples
Experimental program
Comparison scheme
Effect test
example 1、2
[0071]
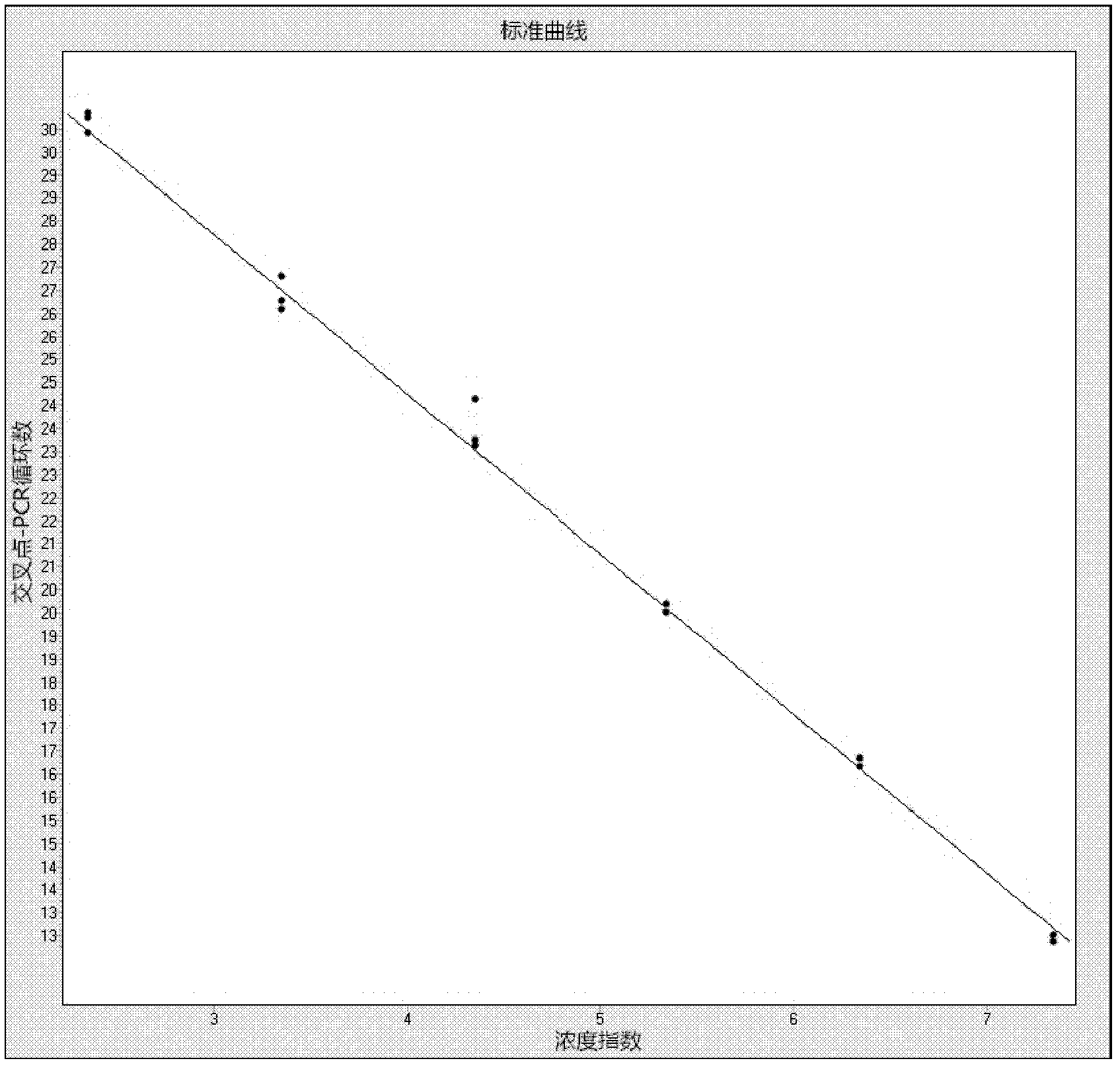
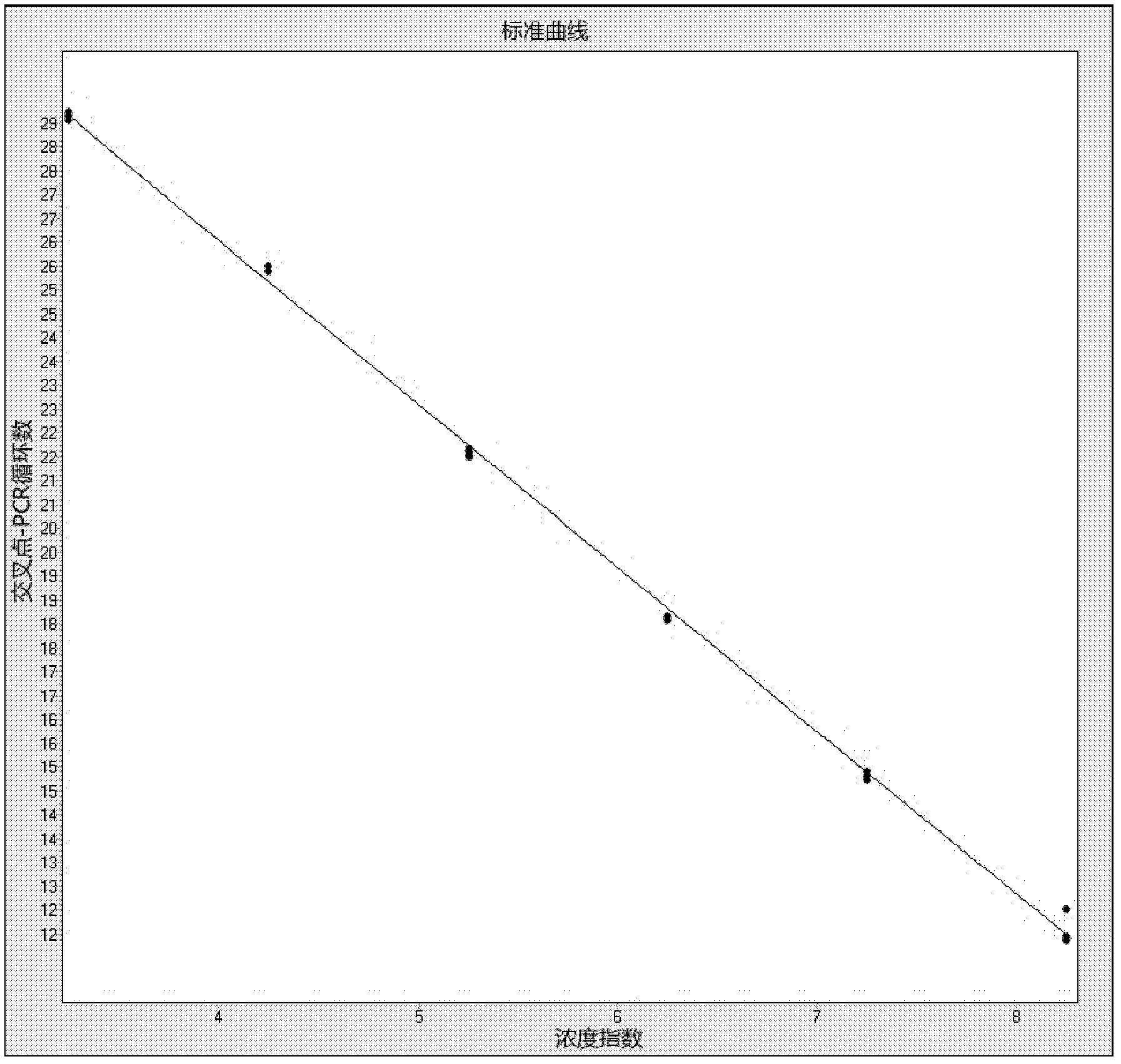
[0072] Eco and Bif single detection and multiple detection were performed on the fecal bacterial nucleic acid of two clinical samples No. 24 and No. 26, and the two detection methods were compared. The number of bifidobacteria and Escherichia coli detected by the two detection methods was very similar.
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
Login to View More PUM
| Property | Measurement | Unit |
|---|---|---|
| PCR efficiency | aaaaa | aaaaa |
Login to View More
Abstract
The invention discloses a method for real-time quantitative polymerase chain reaction (PCR) detection of bifidobacteria and Escherichia coli by using Taqman probes. The method comprises the following steps of: A) extracting bacterial genome DNA of the bifidobacteria and the Escherichia coli in a sample to be detected; B) synthesizing primers and the Taqman probes according to related sequences of the bifidobacteria and the Escherichia coli; and C) ensuring that the primers and the probes obtained in the step B) and other PCR reaction reagents form a reaction system for real-time quantitative PCR by the Taqman probes. The Taqman probes are labeled by double colors, by a double absolute quantitative PCR method, two kinds of intestinal bacteria in a sample can be accurately quantified at one time, the detection cost and detection time can be saved, and errors caused by repeated detection are reduced.
Description
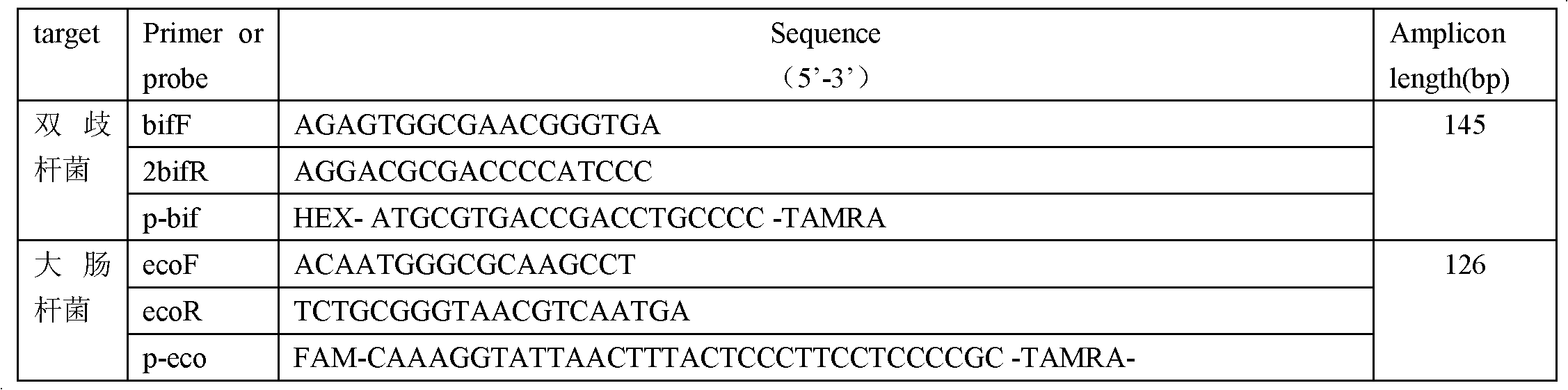
technical field [0001] The invention relates to the field of molecular biology detection, in particular to a method for detecting bifidobacteria and Escherichia coli by using Taqman probes and multiple primers in real-time quantitative PCR. Background technique [0002] In the current study of stool samples from healthy people, it is known that about 10 14 There are about 400 to 500 species of bacteria, which are 10 to 20 times the total number of human cells. A large number of research results show that the bacteria in the intestinal tract play an important role in the growth and development of the human body, immune function, nutrition, pathogenesis of various diseases and health. Therefore, the quantification of intestinal bacteria is very important for studying the function of intestinal flora and the relationship with the body. [0003] When detecting bacteria, the most traditional method is to use traditional culture counting, but because there are a large number of ...
Claims
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
Login to View More Application Information
Patent Timeline
 Login to View More
Login to View More Patent Type & Authority Applications(China)
IPC IPC(8): C12Q1/68C12Q1/04C12R1/19C12R1/01
Inventor 马卓娅郑跃杰
Owner SHENZHEN CHILDRENS HOSPITAL
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com