Detection method of fetal chromosomal aneuploidy
A technology of aneuploidy and chromosome, which is applied in the field of medical testing, can solve problems such as misjudgment and reduce the reliability of testing, and achieve the effects of improving accuracy, reducing waste, and increasing the scope of application
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
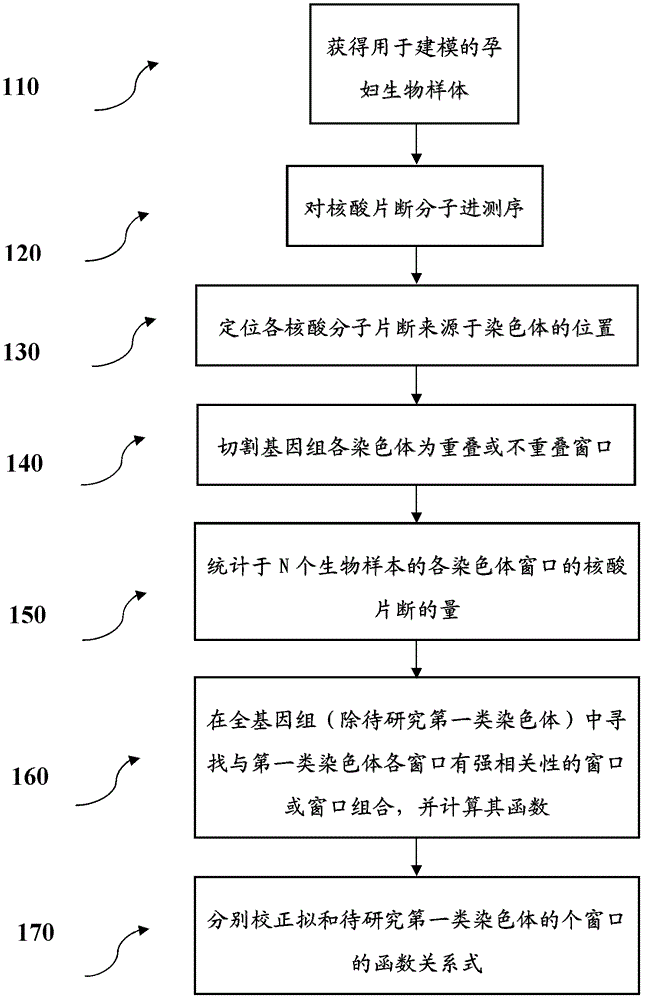
[0075] Step 1: Obtain the sequencing data required for modeling
[0076] 89 normal samples were selected, and these samples were prepared and sequenced under different experimental conditions.
[0077] Briefly, the DNA library is prepared first, and then the nucleic acid fragment molecules are ligated to the Solexa sequencing adapter, and then the nucleic acid molecules with a nucleic acid fragment length of 150 to 300 base pairs are isolated and purified. The nucleic acid molecules connected with the adapter can hybridize with the complementary adapter on the surface of the flow cell. Under certain conditions, the nucleic acid molecules grow in clusters, and then go through 36 rounds of sequencing cycles on the Illumina Genome Analyzer, which is equivalent to measuring 35 bases per nucleic acid molecule. base-pair nucleic acid fragments. The sequence information measured next goes through a pipeline process, and finally the ELAND comparison result with the human genome seque...
Embodiment 2
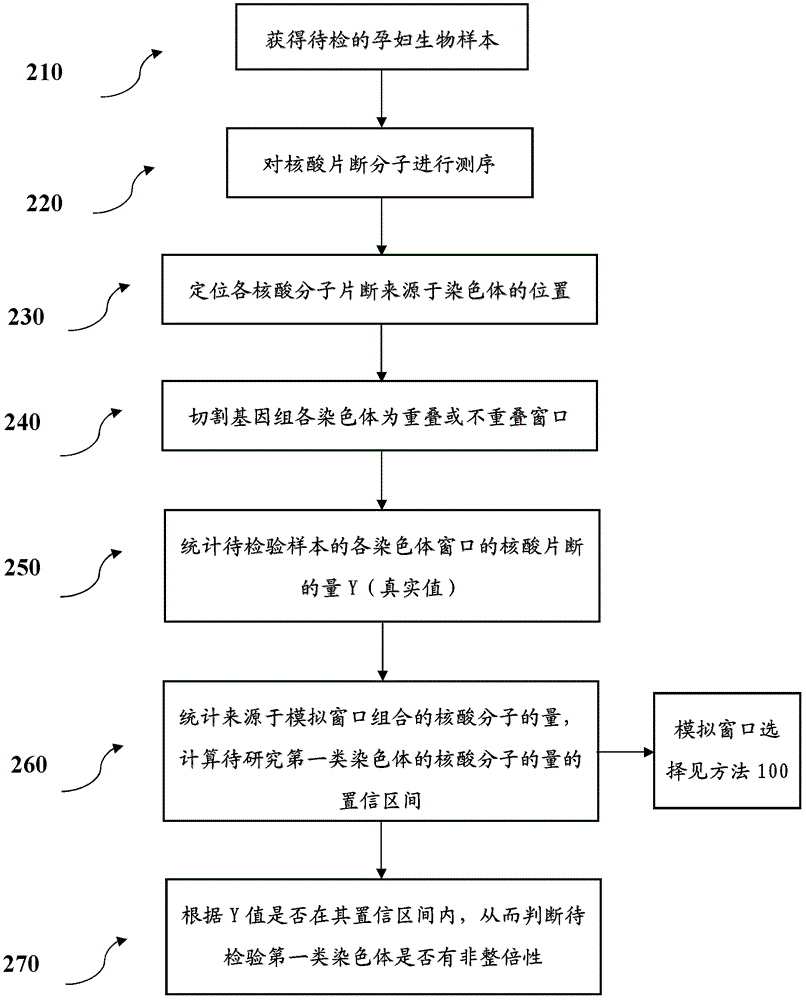
[0124] Step 1: Obtain the sequencing data required for modeling
[0125] 53 normal samples were selected, and these samples were prepared and sequenced under different experimental conditions.
[0126] This step corresponds to figure 1 110, 120 steps.
[0127] Step 2: Build a detection model for chromosome 21
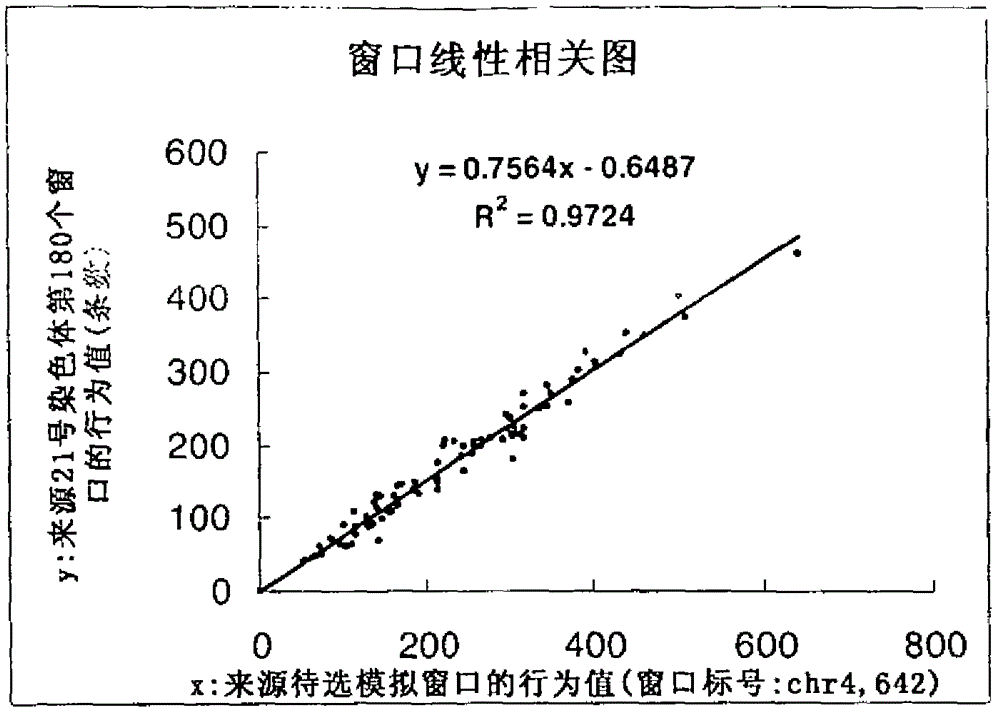
[0128] according to figure 1 The flow chart is carried out, and the process of establishing the mathematical model is optimized by the computer program. The computer program completes the search of the correlation window through different parameter selections, finds the optimal parameters, and establishes the functional relationship between the first type of chromosome window and the second type of chromosome window.
[0129] In step 130, based on sequencing, each nucleic acid fragment molecule measured can be positioned to the exact position of the genome by means of biological information analysis.
[0130] In step 140, a window is cut for each sample. Each chro...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com