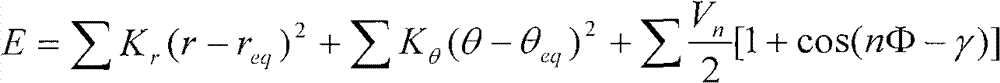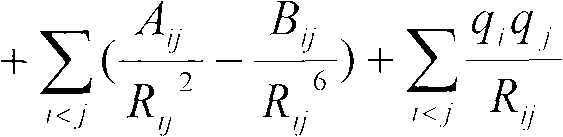Method for establishing molecular simulation force filed of protein system
A molecular simulation and protein technology, applied in special data processing applications, instruments, electrical digital data processing, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0015] Using the amino acid tetrapeptide chain structure (ACE-ALA-XXX-ALA-NME, XXX=one of 20 amino acids) as a model, we selected five representative structures (right-helical (α R , φ=-57.0, ψ=-47.0), left helical (α L , φ=57.0, ψ=47.0), β-sheet (β, φ=-119.0, ψ=113.0), antiparallel β-sheet (β a , φ=-140.0, ψ=135.0), PPII (PPII, φ=-79.0, ψ=150.0), the selection of the side chain dihedral angle is the same as that of the dipeptide model side chain dihedral angle), and MP2 / cc - The result of pVTZ calculation is used as a standard to check the accuracy of the force field. In order to compare the relative energies of the five configurations of the 20 amino acids with different quantum chemistry and other versions of the AMBER force field method. In the calculation of the density functional method M052x and B3LYP, the configuration of the model molecule is firstly optimized in the gas state using the 6-31G** basis set, and the ASP and GLU models are optimized using the 6-31+G* ba...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com