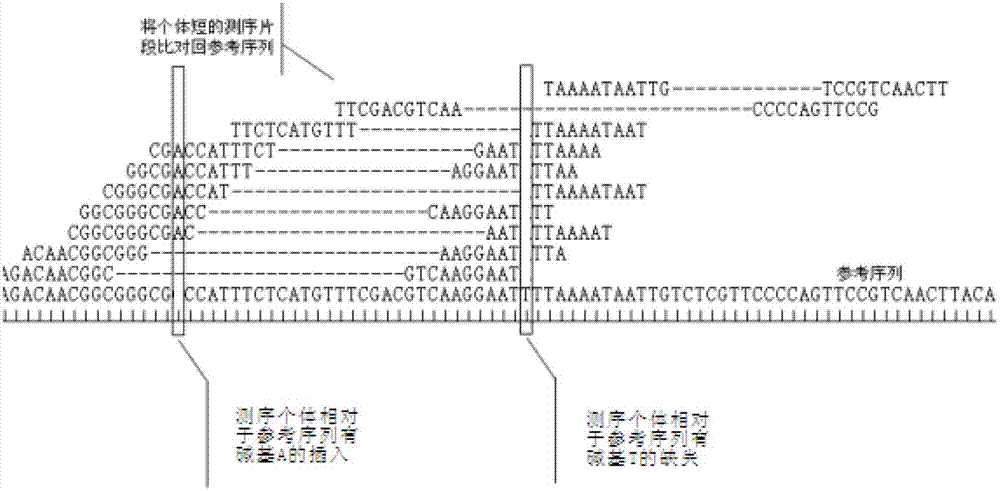
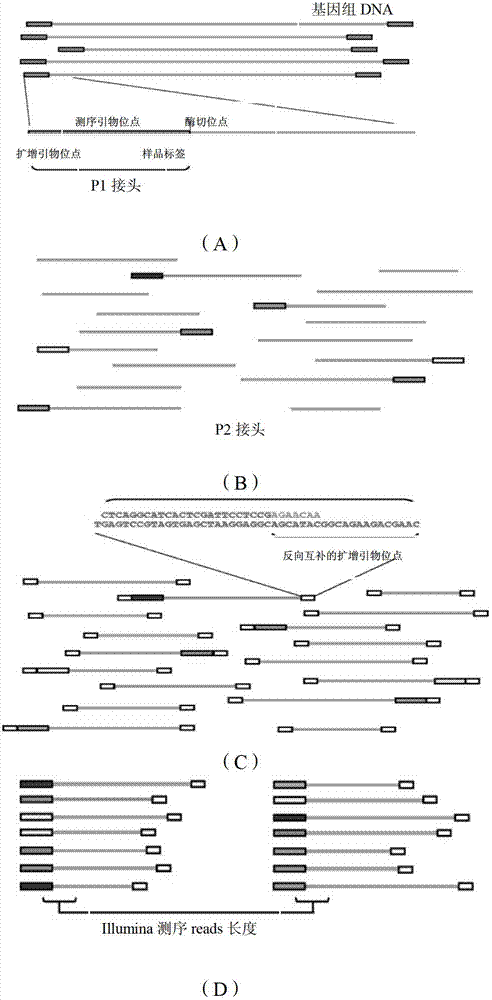
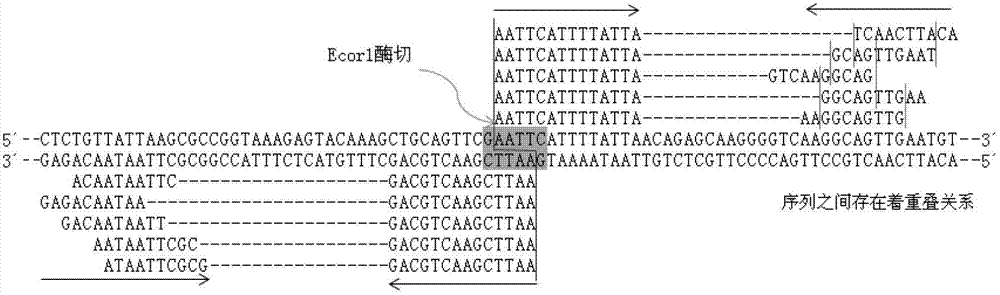
Primer design developing method of length polymorphism sign based on restriction enzyme digestion database-establishing pair-end sequencing
A paired-end sequencing and length polymorphism technology, applied in the field of bioinformatics, can solve the bottleneck of Indel labeling technology and the absence of reference sequences, etc., and achieve the effect of reducing sequencing costs and simplifying complexity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0063] Embodiment specific operation process:
[0064] The sequencing data obtained by the RAD-PE sequencing of the two parents were filtered according to the sequencing quality value, N content, and whether it contained restriction end sequences, and unqualified sequencing sequences were removed. The effective data statistics obtained are shown in Table 1.
[0065]
[0066] Table 1. Effective data statistics of gourd RAD-PE sequencing
[0067] According to the sequencing sequence of the enzyme-cleaved end of the sequenced individual genome, the information of the two parental piles is respectively generated by using the identity of the sequence. Filter out the results with a sequencing depth of 1 for the restriction end sequence. Figure 10 The distribution map of RAD sequence sequencing depth is shown, and the statistics of the results are shown in Table 2.
[0068] gourd sample Number of RAD sequences Average sequencing depth of RAD sequences male pare...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com