Single nucleotide polymorphism for prognosis of hepatocellular carcinoma
A single nucleotide polymorphism, hepatocellular carcinoma technology, applied in the field of single nucleotide polymorphism, can solve the problems of low survival rate and high relapse rate, and achieve the effect of improving poor prognosis
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0053] Embodiment 1 SNP selection
[0054] Biallelic SNPs contained within ±2 kb of angiogenesis-related genes (ie, VEGF, HIF1a, IGF2, FGF2 or MTA1) were investigated. For the position of the SNP of the gene, refer to the known gene information (http: / / www.ncbi.nlm.nih.gov / project / SNP) for the 5'-untranslated region, promoter region, exon region and gene Seating area to choose from.
[0055] The IDs of the SNPs under study are as follows:
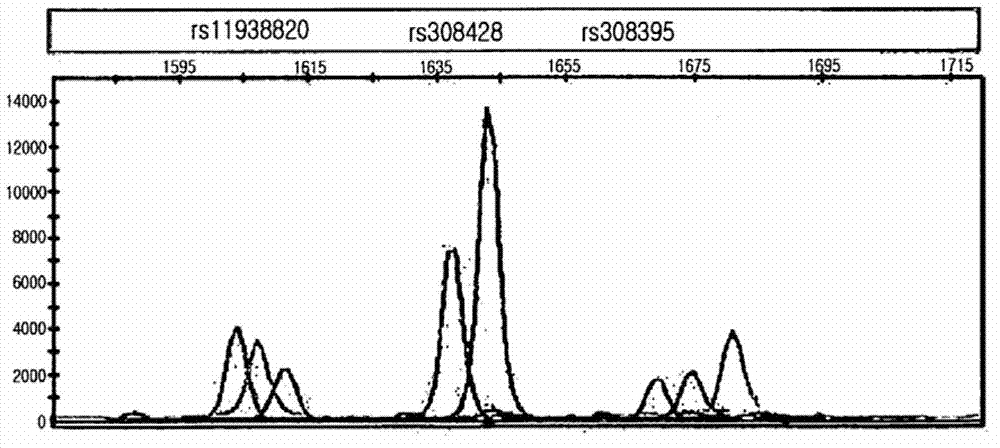
[0056] rs699947、rs25648、rs3025000、rs3025010、rs3025035、rs3025040、rs10434、rs998584、rs45533131 / rs1957757、rs2301113、rs2057482 / rs2585、rs3802971、rs3213221、rs3741212、rs2239681、rs3741208、rs1004446、rs7924316、rs3842748、rs2070762 / rs308395、rs308428、rs11938826、 rs17472986, rs308442, rs308379, rs308381, rs6534367, rs1048201, rs3747676 / rs4983413 and DL1002505.
[0057] Among the above SNPs, SNPs with a haplotype frequency equal to or higher than 5% were selected, and the haplotype frequencies were analyzed by using PHASE software v2.1. In addition, li...
Embodiment 2
[0058] Embodiment 2 SNP genotype analysis
[0059] 1. SNP genotype analysis
[0060] A primer set capable of amplifying the region containing the SNP of Example 1 and a TaqMan probe containing the SNP region were designed by using the primer express software. For TaqMan probes, individual probes for wild-type and mutant alleles were designed according to the sequence of the SNP.
[0061] Probes are made by adding a fluorochrome to one side of a TaqMan probe and a quencher that suppresses the color of the fluorochrome on the other side. In this case, each fluorescent dye with a different color is added for the wild-type and mutant alleles, respectively.
[0062] The three types of primers disclosed in Table 1 below were mixed together, and then a PCR reaction was performed by using the mixed primers, thereby discriminating single nucleotide pair mismatches according to the exonuclease activity property of Taq polymerase.
[0063] [Table 1]
[0064]
[0065]
[0066] A...
Embodiment 3
[0070] Example 3 patient characteristics
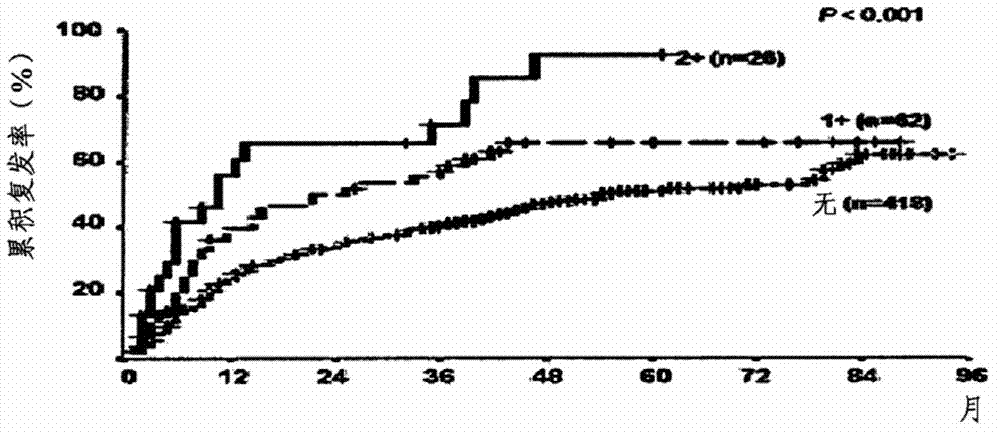
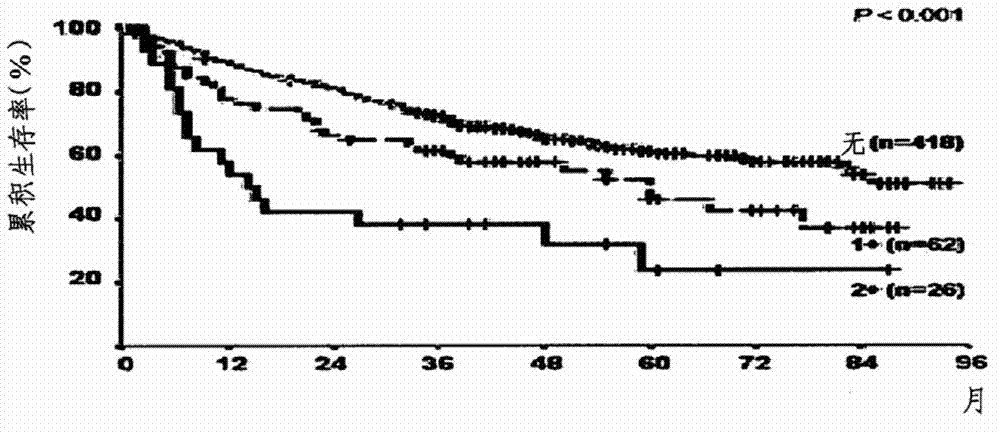
[0071] The experiment was conducted on 506 patients with recurrent HCC at Asan Hospital between 1998 and 2003. The clinical characteristics of the 506 patients are shown in Table 2 below. Following radical surgical resection, patients were followed at a mean interval of 43 months (range, 1-96 months).
[0072] [Table 2]
[0073]
[0074] Survival and HCC recurrence were determined by using medical records from the last day of follow-up. For patients who have not been followed up for 3 months, the evaluation is carried out by visiting the nearby residents of the patient.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com