Method for rapidly discriminating species of cotton
A cotton and cotton seed technology, applied in the field of molecular biology, to achieve the effect of simplifying the identification process
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1 20
[0024] The acquisition of the DNA bar code of twenty known cotton species of embodiment 1
[0025] 1 Materials and methods
[0026] 1.1 Experimental materials
[0027] The experimental materials are the leaves of 20 known cotton species, numbered 1-20 respectively, from the Cotton Research Institute of the Chinese Academy of Agricultural Sciences, as shown in the table below:
[0028]
[0029] 1.2 Extraction of total DNA
[0030] The total DNA of 20 cotton species was extracted by CTAB method. Purified DNA was tested for its purity and concentration by UV spectrophotometer, and its quality was tested by 1% agarose gel electrophoresis. The DNA concentration was diluted to about 60ng / μl, and stored in a 4°C refrigerator for use as a working solution.
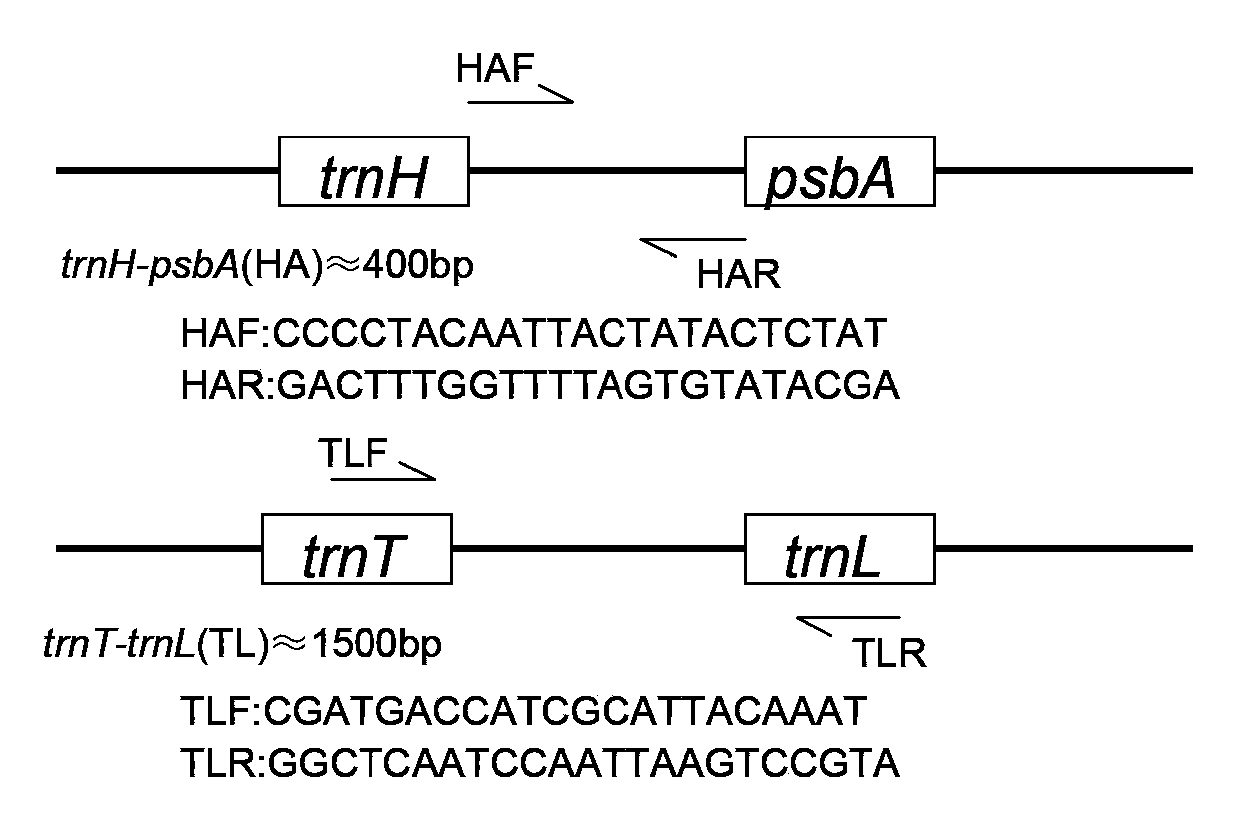
[0031] 1.3PCR amplification
[0032] Design landing PCR and use universal primers for amplification: the universal primer pair includes TL and HA. For trnT-trnL, the nucleotide sequence of the upstream primer TLF is: CGATG...
Embodiment 2
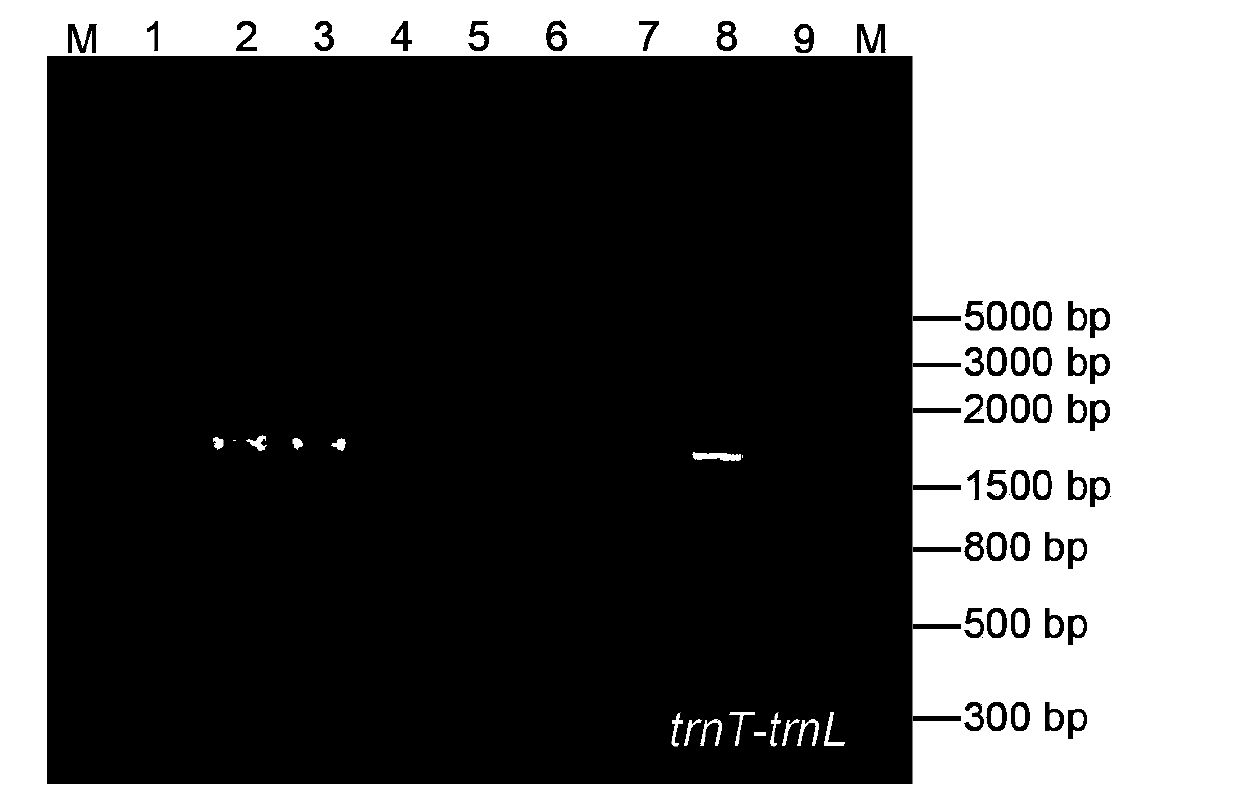
[0044] Example 2 DNA barcode identification technology of unknown cotton species
[0045] 1 Materials and methods
[0046] 1.1 Experimental materials
[0047] The experimental materials are the leaves of nine different cotton species, numbered 1-9 respectively, from the Cotton Research Institute of the Chinese Academy of Agricultural Sciences.
[0048] 1.2 Extraction of total DNA
[0049] The total DNA of nine cotton species was extracted by CTAB method. Purified DNA was tested for its purity and concentration by UV spectrophotometer, and its quality was tested by 1% agarose gel electrophoresis. The DNA concentration was diluted to about 60ng / μl, and stored in a 4°C refrigerator for use as a working solution.
[0050] 1.3PCR amplification
[0051] Design landing PCR and use universal primers for amplification: the universal primer pair includes TL and HA. For trnT-trnL, the nucleotide sequence of the upstream primer TLF is: CGATGACCATCGCATTACAAAT, and the nucleotide seque...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com