Method for realizing parallel compression and parallel decompression on FASTQ file containing DNA (deoxyribonucleic acid) sequence read data
A decompression and data technology, applied in concurrent instruction execution, electrical digital data processing, special data processing applications, etc., can solve problems such as parallel algorithm research articles that have not yet seen multi-core CPUs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
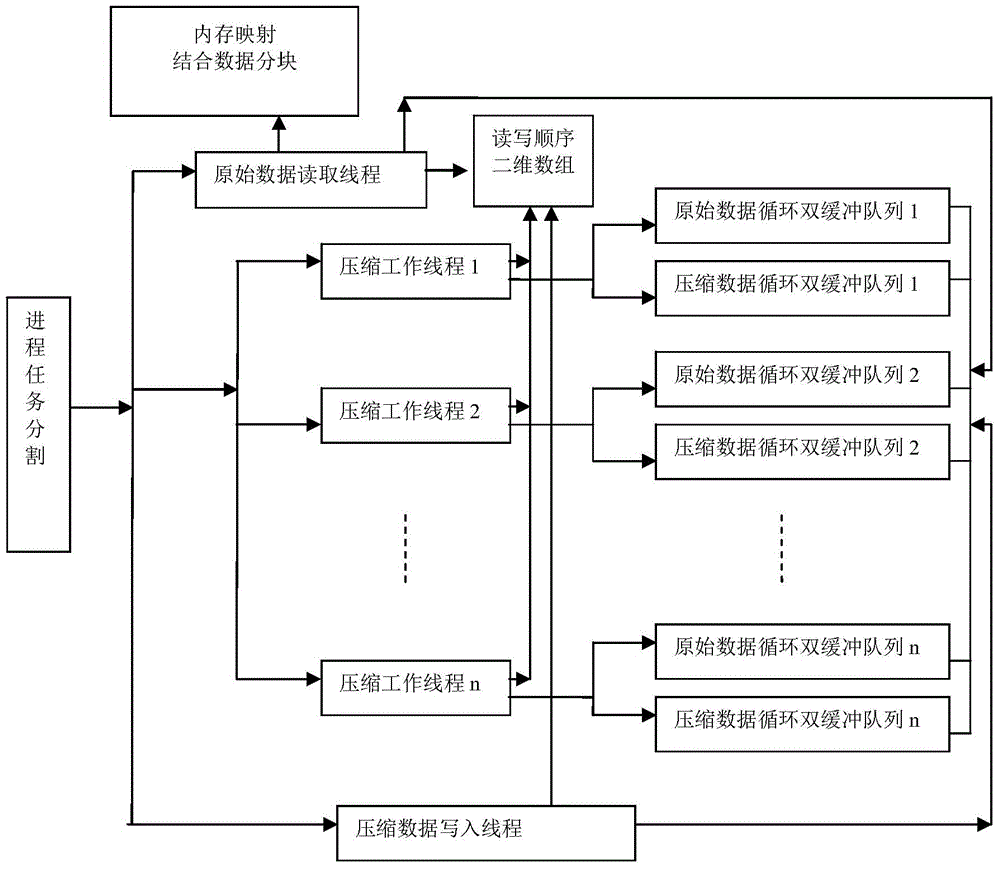
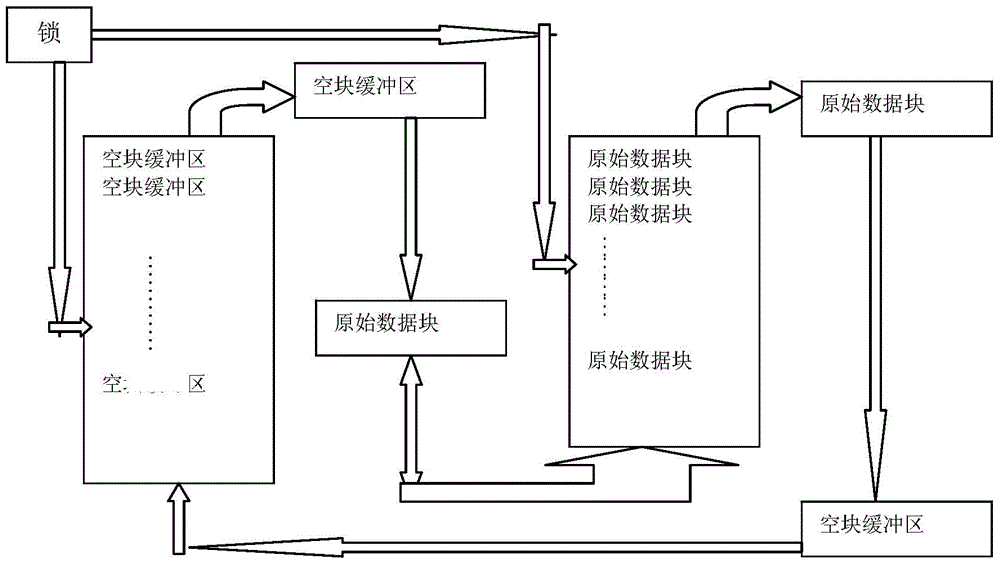
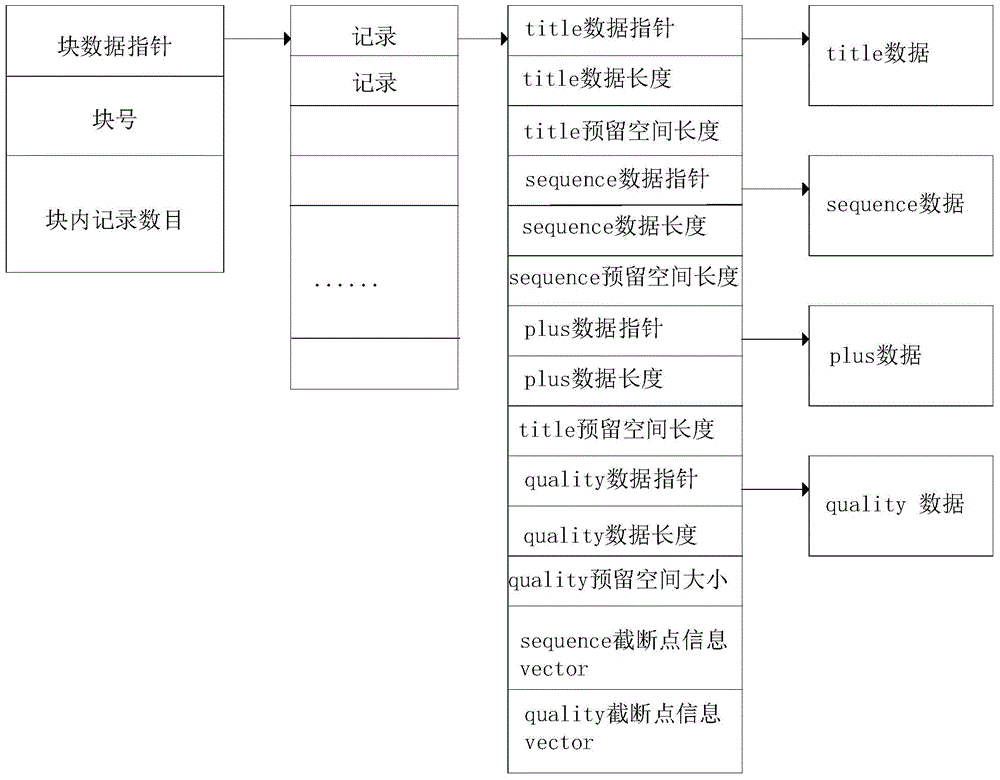
[0062] The present invention provides a method for parallel compression and decompression of FASTQ files of DNA reading sequence data. In order to make the purpose, technical solution and effect of the present invention clearer and clearer, the present invention will be further described in detail below in conjunction with the accompanying drawings. It should be understood that the specific embodiments described here are only used to explain the present invention, not to limit the present invention.
[0063] The raw data reading thread in the parallel compression method of the FASTQ file is explained in detail below, and its specific implementation steps are as follows:
[0064] (1) Open the FASTQ compressed file of the raw DNA read sequence data to be compressed.
[0065] (2) Obtain the memory paging size of the file system of the currently running machine.
[0066] (3) Set the memory mapping space size according to the memory paging size.
[0067] (4) According to the rang...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com