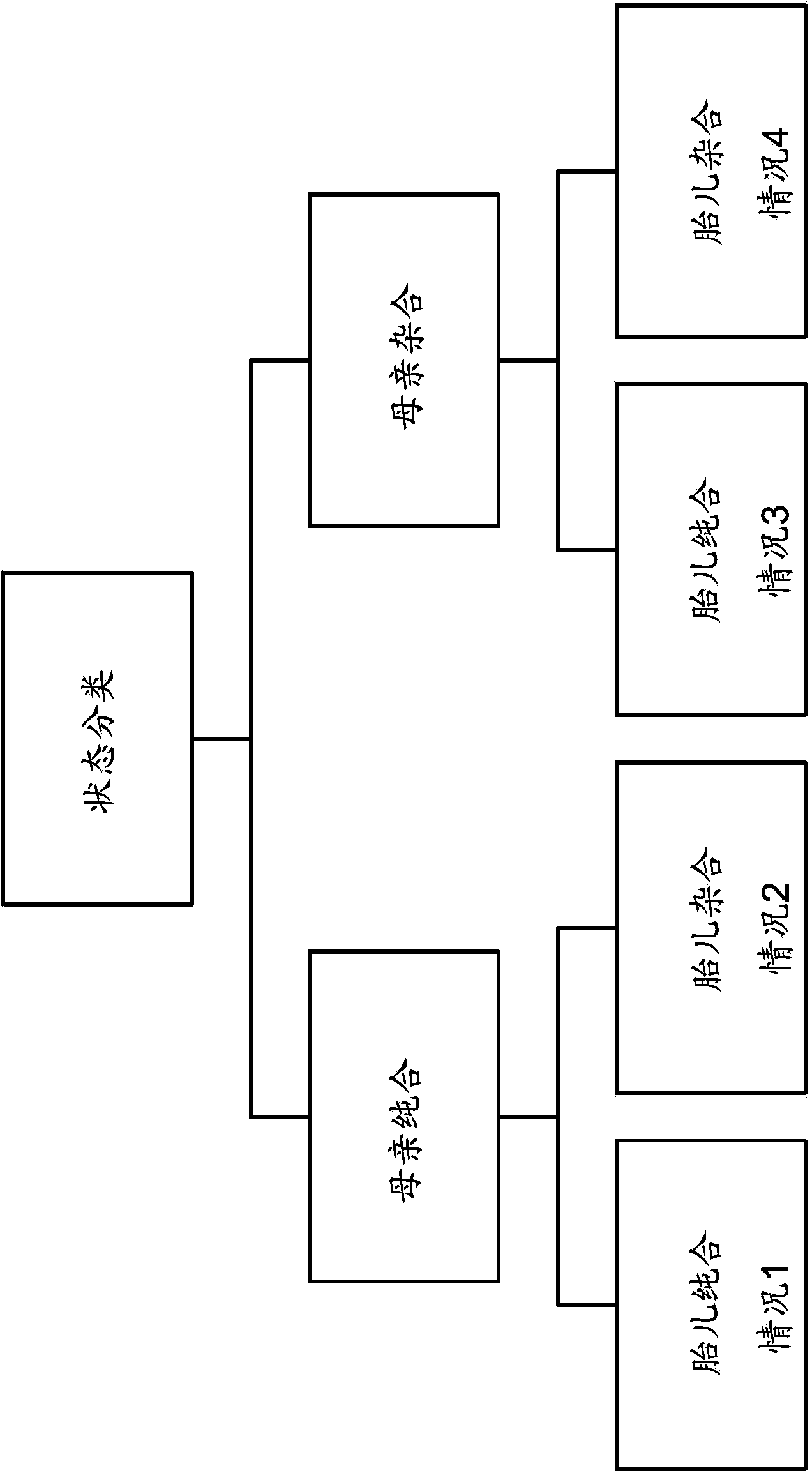
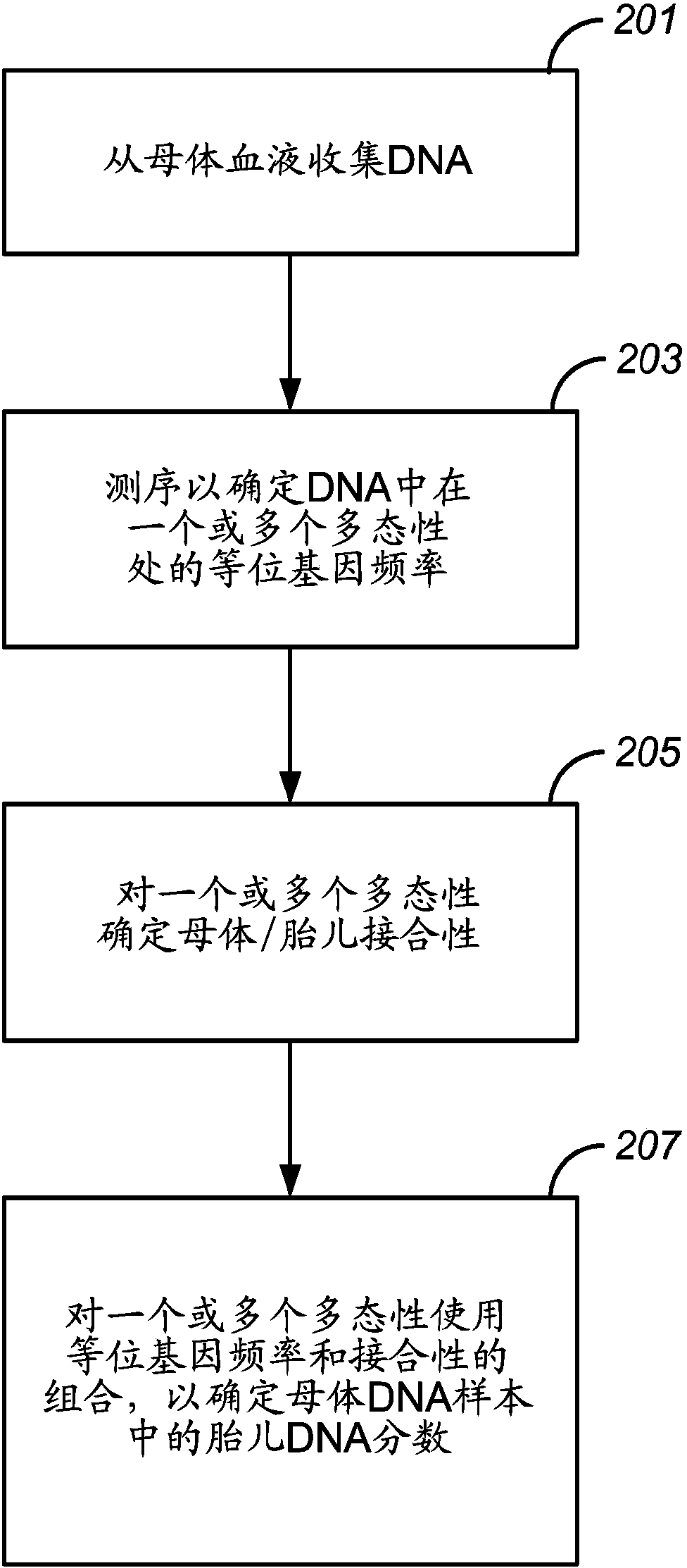
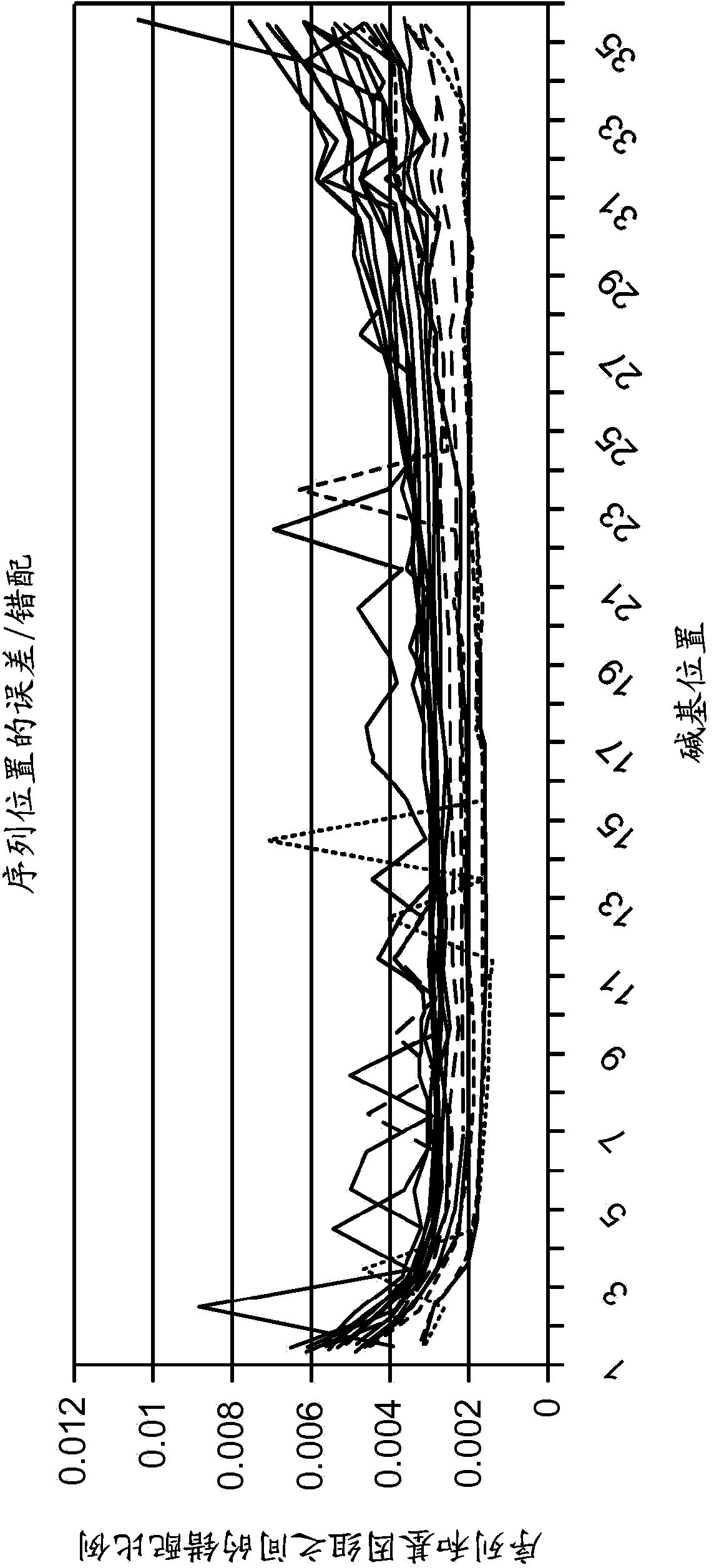
Resolving genome fractions using polymorphism counts
A genomics, fractional technology, used in genomics, sequence analysis, proteomics, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0054] Introduction and Overview
[0055] Certain disclosed embodiments relate to analyzing DNA obtained from the blood of a pregnant female, and using the analysis to determine the fraction of that DNA from the fetus. The fetal fraction of DNA can then be used to attribute a certain level of confidence to another measure or characterization of the fetus based on a separate analysis of DNA obtained from the mother's blood. For example, a fetal DNA sample obtained from maternal blood can be analyzed separately to detect aneuploidy in a fetus conceived by a pregnant woman. The determination of aneuploidy by this separate analysis can be given by a statistically robust confidence level based on the fractional amount of fetal DNA present in the DNA obtained from the maternal blood. A lower fraction of fetal DNA in the total complement of DNA indicates low confidence in any characterization based on fetal DNA.
[0056] Typically, but not necessarily, the analyzed DNA in materna...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com