Fluorescence labeling multiplex PCR detection kit for Clostridium difficile
A detection kit and technology for Clostridium difficile, which are applied in the direction of microbial determination/inspection, biochemical equipment and methods, etc., can solve the problems of low sensitivity of quantitative PCR, no multiple PCR amplification, etc., to prevent misjudgment. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] Example 1 Design of the kit
[0042] Download the primer design template sequence from NCBI genebank, as shown in Table 4.
[0043] Table 4 Template sequence list
[0044] Locus
Sequence Genebank Number
Location
tpi
AM180355.1
3706953-3707696
tcdA
JQ809335.1
1-8133
tcdB
AF217292.1
1-7512
ctdC
HQ596359.1
1-1006
cdtA
AF271719.1
1-1392
cdtB
AF271719.1
1445-4075
[0045] The sequence of each locus is aligned in genebank, and the most conserved region is selected to design primers.
[0046] Taking the primer design of TcdA as an example, the full sequence of the primer design template JQ809335.1 was aligned in NCBI blast, and the sequence specificity of nt4000-6000 was determined to be high. Using this partial sequence as the primer design template, 4 primer sets were designed, as follows table.
[0047] Table 5 Initially designed primers for TcdA
[0048] The primers are synthesized by Shanghai Shenggong, and a single pair of primers are amplified by anneali...
Embodiment 3
[0095] Thirteen stool samples were donated by the Zhejiang Center for Disease Control and Prevention. The strains were identified and analyzed at the Zhejiang Center for Disease Control and Prevention by strain culture method and single-plex quantitative PCR.
[0096] The fecal genome was extracted using Qiagen’s QIAamp DNA stool Mini Kit, according to the operating instructions, and the template was quantified at a concentration of 0.5-2ng / μl.
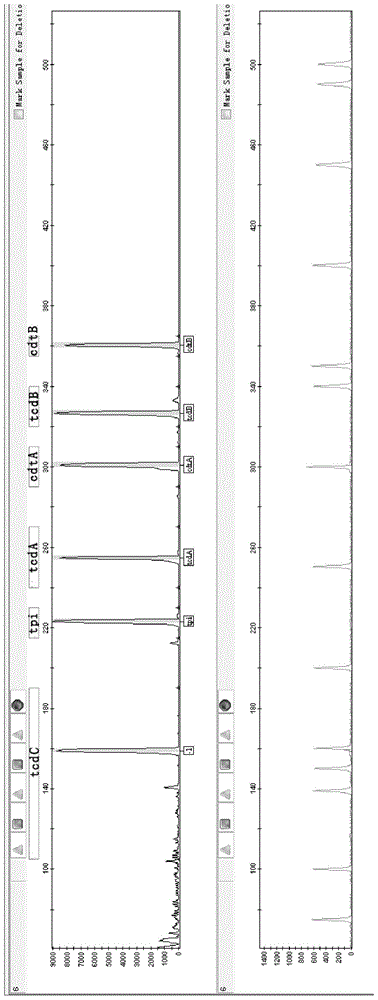
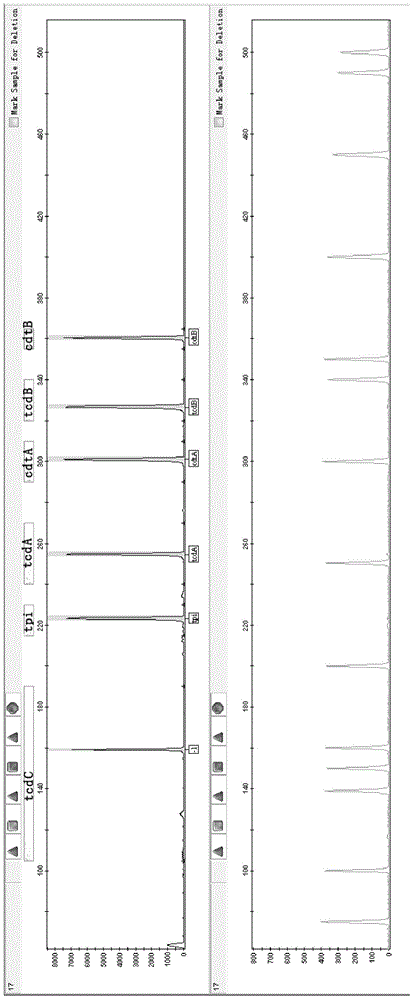
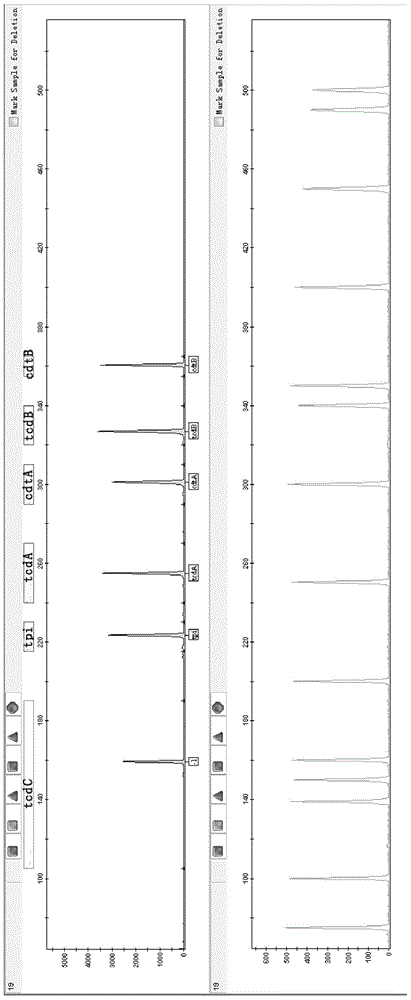
[0097] The 13 fecal genomic DNAs were amplified according to the primer set, amplification system, and amplification program finally determined in Example 1, and tested as in Example 1. The software analyzes that the peak height is higher than 200 rfu, and it is considered that the corresponding gene has been amplified. On the contrary, if the peak height of the analysis result is less than 200rfu, it is considered that there is no amplification, that is, it does not contain the corresponding gene.
[0098] The 13 samples were classified in...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com