Method for preparing length relying probe for detecting gene mutation
A length-dependent, long-probe technology, applied in the field of oligonucleotide probe preparation, can solve the problems of cumbersome biological preparation methods, achieve diverse and convenient sources, simple primers, and improve the detection success rate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
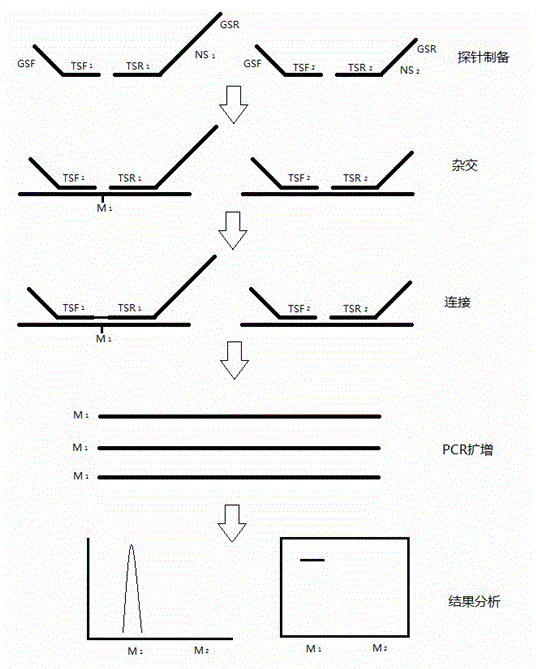
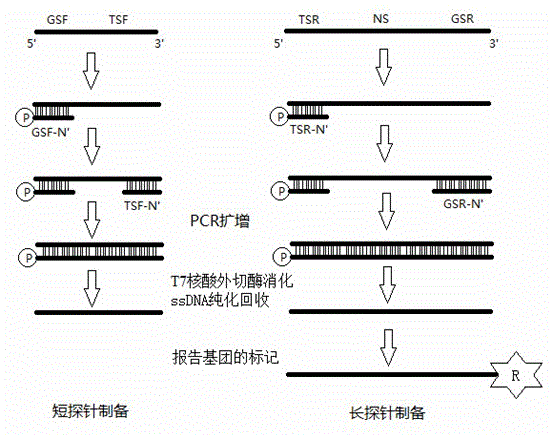
[0046] Example 1 The preparation and detection of the length-dependent probe pair for detecting the mammalian cardiovascular disease-related gene ACE gene includes the following steps:
[0047] 1. Design two pairs of primers for the ACE gene to be tested. One pair is a universal primer for subsequent PCR amplification detection. They are GSF and GSR respectively. The natural sequence (NS) is irrelevant; the detection primers for the target site or fragment to be detected are TSF and TSR, respectively.
[0048] 2. Prepare the long probe in the length-dependent probe pair:
[0049] 1) Determine a natural sequence (NS) that has nothing to do with the ACE gene to be tested and the universal primer, according to the NS and the sequence of the ACE gene to be tested within the chicken house;
[0050] 2) For long probes, a pair of amplification primers are involved, namely GSR-N' and TSR-N', wherein GSR-N' is composed of the sequence of GSR and the sequence of about 18bp at the 3' en...
Embodiment 2
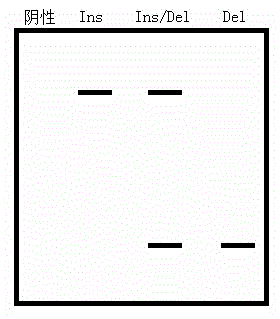
[0152] Example 2 The preparation and detection of length-dependent probe sets for detection of mammalian cardiovascular disease-related genes ACE, GNB3, and NOS3 genes include the following steps:
[0153] 1. Design a pair of universal primers for the length-dependent probe sets of ACE, GNB3, and NOS3 genes for subsequent PCR amplification detection, which are GSF and GSR respectively. The native sequence (NS) in the probe is irrelevant.
[0154] The concrete nucleotide sequence of above-mentioned primer is:
[0155] GSR: 5'-CACACAGGAAACAGCTATGAC-3' 21bp
[0156] TSR: 5'-ACCTGCTGCCTATACAGTCACTTTTT-3' 25bp
[0157] 2. Design a pair of detection primers for the target site or fragment of the ACE gene to be tested, namely TSF and TSR.
[0158] 3. Design a pair of detection primers for the target site or fragment of the GNB3 gene to be tested, namely TSF1, TSRC1 and TSRT1.
[0159] 4. Design a pair of detection primers for the target site or fragment of the NOS3 gene to be tes...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com