Tandem mass spectrogram identification method
An identification method and tandem mass spectrometry technology, applied in special data processing applications, measuring devices, instruments, etc., can solve the problems of large search space, reduced search speed, and increased number of peptides, and achieve high search speed, improve accuracy, The effect of improving the identification rate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
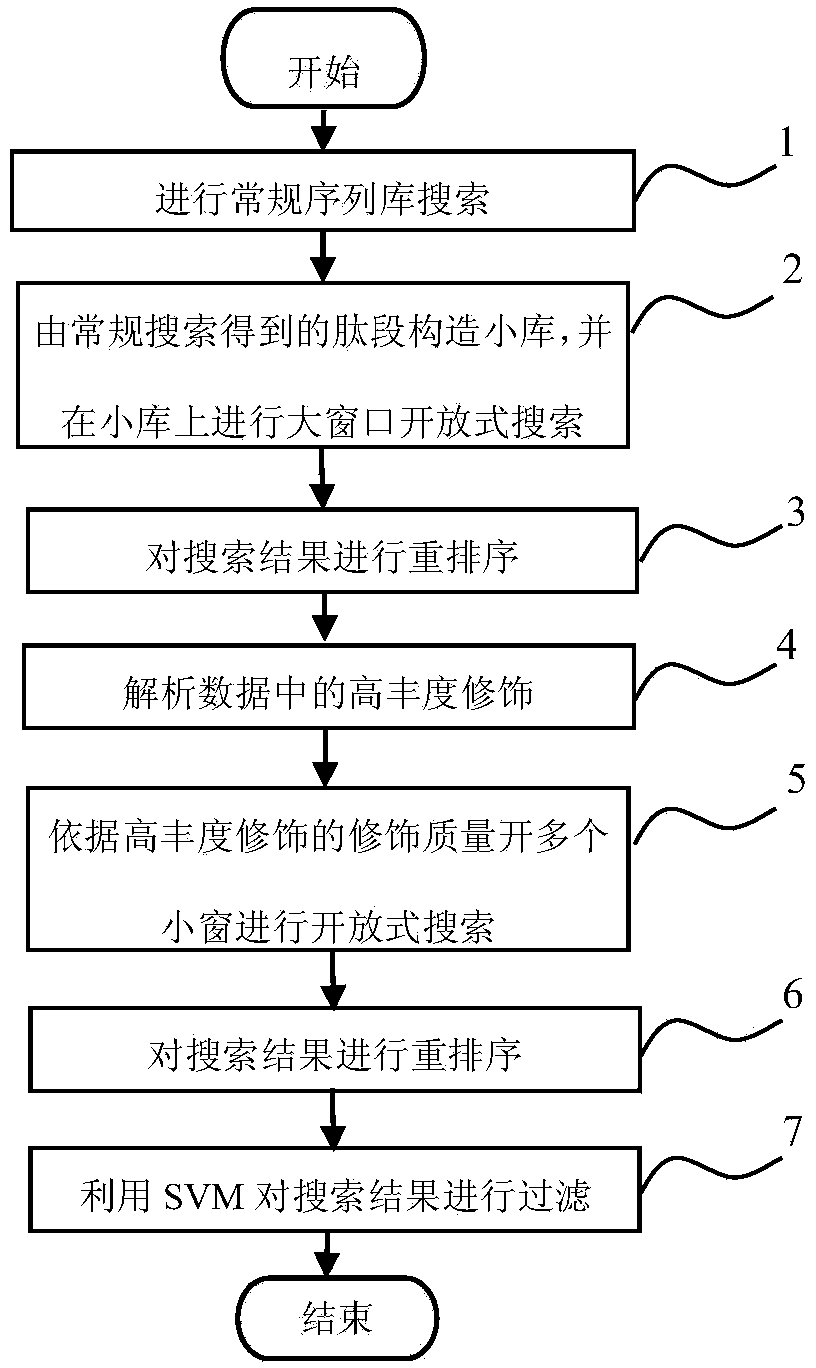
[0031] figure 1 Shows a flow chart of a method for identifying a tandem mass spectrum spectrum according to an embodiment of the present invention. The method for identifying a tandem mass spectrum spectrum includes the following steps:
[0032] Step 1: To identify the tandem mass spectrum data set, for each tandem mass spectrum (tandem mass spectrum is the output signal of the mass spectrometer, for ease of description, hereinafter referred to as the spectrum), respectively, based on the global sequence library Search within a small mass window and identify some peptides. The search in this step is the conventional search on the global sequence library (ie, non-open search, also known as restricted search). The small window refers to the quality interval centered on the quality of the spectrum to be identified, and the quality interval is relatively narrow. For example, if the mass of the spectrum to be identified is m, the corresponding small window is [m-0.00002m, m+0.00002m...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com