Zero-background cloning vector as well as preparation method and application thereof
A cloning vector, zero background technology, applied in the field of bioengineering, can solve problems such as gene inability to express, plasmid interference, and vector quality influence
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1 0
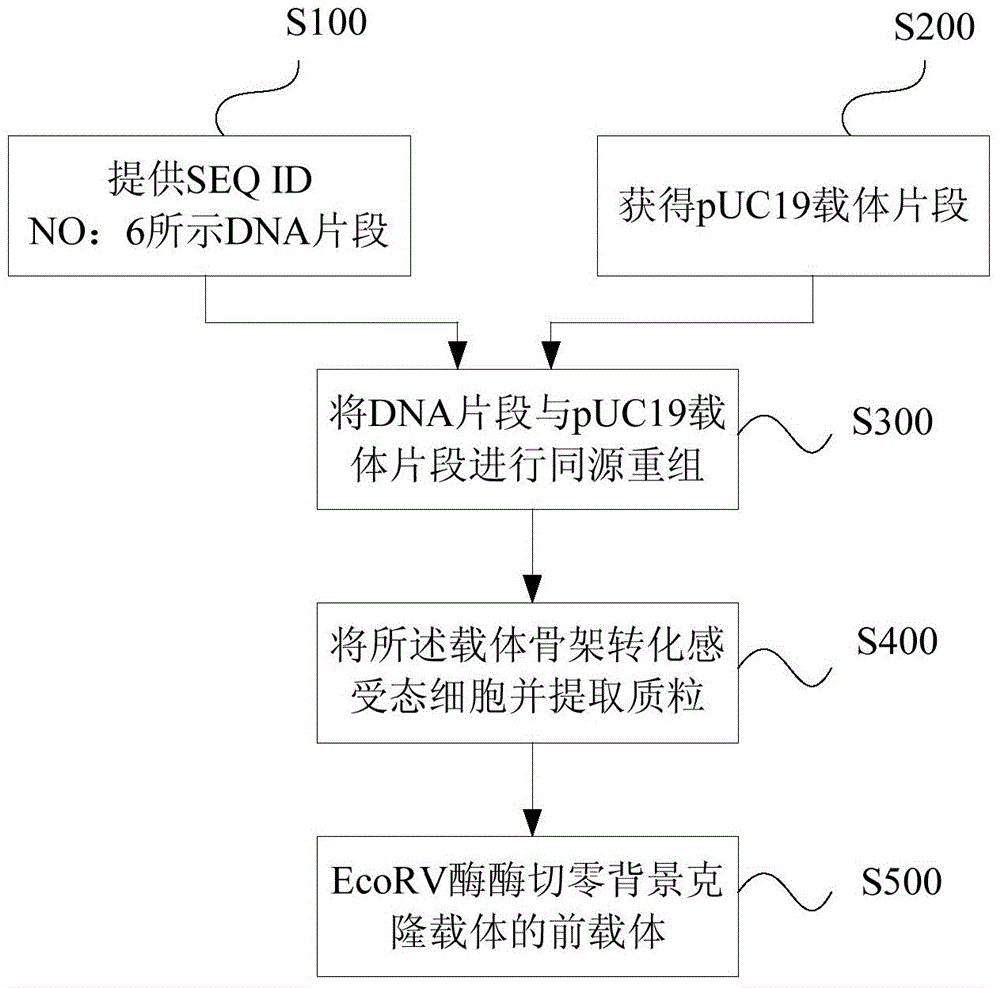
[0086] Example 1 Preparation of zero background cloning vector
[0087] To prepare a zero-background cloning vector, it is first necessary to construct a pre-vector of a zero-background cloning vector, and the inventors selected a high-copy pUC19 plasmid for transformation. Proceed as follows:
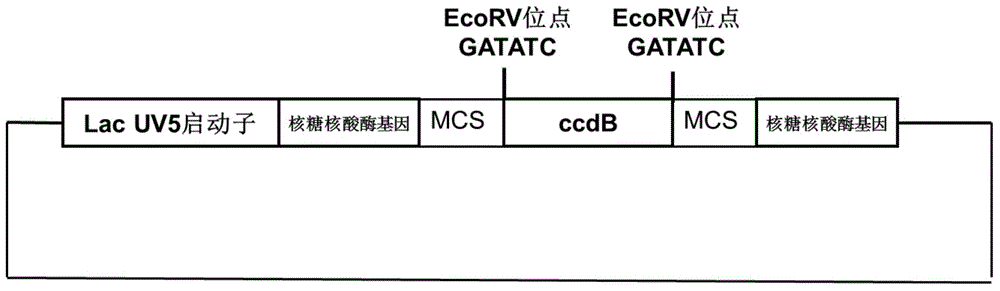
[0088] (1) Artificially synthesize a gene sequence shown in SEQ ID NO: 6, which includes the ribonuclease gene driven by the LacUV5 promoter (SEQ ID NO: 4), the spacer sequence of the ccdB gene (SEQ ID NO: 5), and Multiple cloning site (SEQ ID NO:3). Synthetic primers are as follows:
[0089]
[0090]
[0091] (2) Dilute all the primers in (1) above to a concentration of 50 micromole / liter with sterile water, draw 5 microliters of each primer and add it to a PCR tube to form a primer mixture, and then use Fast HiFidelity PCR Kit (Tiangen Biochemical Technology (Beijing) Co., Ltd., article number: KP202), using fast high-fidelity polymerase for the first round of amplification ...
Embodiment 2
[0115] Embodiment 2 uses the implementation method of the connection gene fragment of zero background cloning vector
[0116] 1. Obtaining the target gene fragment
[0117] Using lambda phage DNA as a template, design primers as follows:
[0118] The forward primer of the 700-base gene fragment (hereinafter referred to as 700-F): GAGGGCAAGTATCGTTTCCA (SEQ ID NO: 70)
[0119] The reverse primer of the 700-base gene fragment (hereinafter referred to as 700-R): ACTGGAAAGCAACGAAGTCC (SEQ ID NO: 71)
[0120] The forward primer of the 2000-base gene fragment (hereinafter referred to as 2K-F): ATCTGCCTTTACGGGGATTT (SEQ ID NO: 72)
[0121] The reverse primer of the 2000-base gene fragment (hereinafter referred to as 2K-R): GTACAGCCAAAGGCATCCAT (SEQ ID NO: 73)
[0122] The forward primer of the 8000-base gene fragment (hereinafter referred to as 8K-F): ATCCCATGTCGGCAAGCATAAGC (SEQ ID NO: 74)
[0123] The reverse primer of the 8000-base gene fragment (hereinafter referred to as 8K-R...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com