SgRNA (small guide ribonucleic acid) pair for specifically identifying porcine H11 locus, and coding DNA (deoxyribonucleic acid) and application thereof
A technology for specific identification and DNA molecule application in sgRNA and its coding DNA and application fields, to achieve the effect of reducing off-target phenomenon, strong specificity and increasing efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0024] Example 1 Construction of sgRNA expression plasmid pair
[0025] 1. sgRNA target design
[0026] According to the H11 site of the mouse, find the Eif4 and Drg genes of the pig (the site of the mouse is located in the middle of the two genes), call out the middle region in NCBI to find the H11 site of the pig, and its nucleotide sequence is shown in the sequence table As shown in sequence 5 in,
[0027] Select the sgRNA target for gene knockout according to the PAM sequence, the PAM sequence is NGG:
[0028] sgRNA-L target site position 1 (named H11-sgL2):
[0029] AGATCAGGGTGGGCAGCTCTGGG
[0030] The nucleotide sequence for recognizing the target in the corresponding sgRNA-L sequence is shown in sequence 1 in the sequence listing; the DNA sequence encoding the above sequence is shown in sequence 3 in the sequence listing.
[0031] sgRNA-R target site position 2 (named H11-sgR1): TTCCAGGAACATAAGAAAGTAGG, the corresponding sgRNA sequence recognizes the target nucleoti...
Embodiment 2
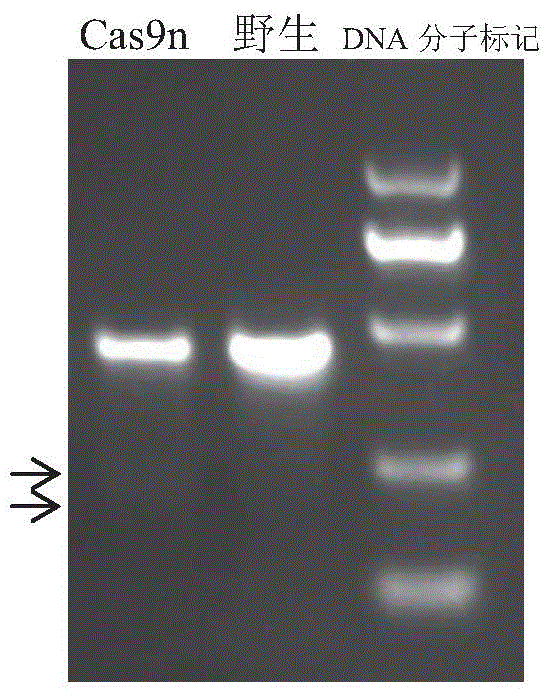
[0041] Example 2 Efficiency verification of sgRNA expression plasmid pair
[0042] 1. Isolation of porcine fetal fibroblasts.
[0043] PEF cells were isolated from aborted pig fetuses. For specific operations, please refer to the literature: Li Hong, Wei Hongjiang, Xu Chengsheng, Wang Xia, Qing Yubo, Zeng Yangzhi; The establishment and biological academic features.
[0044] 2. Nucleofection
[0045] 2 μg each of the recombinant plasmids pX335-sgRNA-H11-L and pX335-sgRNA-H11-R were co-transfected into PEF cells by electroporation to obtain recombinant cells. The specific steps of transfection are as follows: use a nuclear transfer instrument (Amaxa, model: AAD-1001S) and a matching mammalian fibroblast transfection kit (Amaxa, product number: VPI-1002) for transfection. First, use 0.1% trypsin (Gibco, catalog number: 610-5300AG) to digest adherent cells, stop digestion with fetal bovine serum (Gibco, catalog number: 16000-044), and wash with phosphate buffer saline (Gibco, c...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com